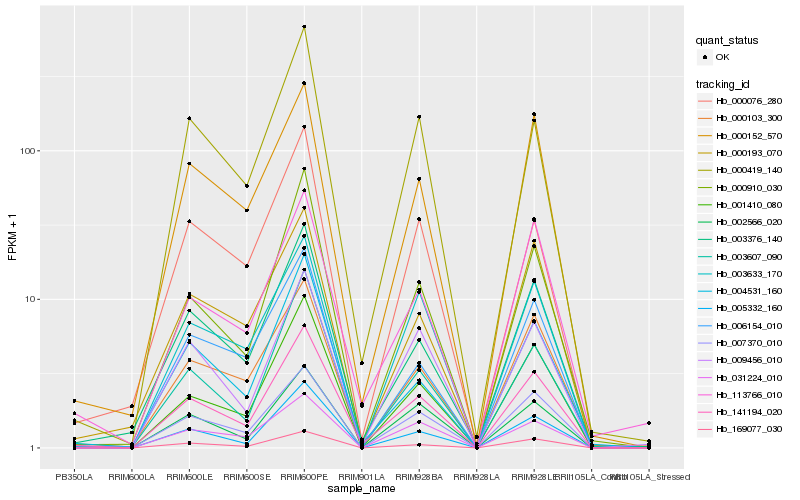
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_141194_020 |
0.0 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 2 |
Hb_001410_080 |
0.0795516727 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_005332_160 |
0.0875002269 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 4 |
Hb_003376_140 |
0.0901942614 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 5 |
Hb_169077_030 |
0.0912603622 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 6 |
Hb_007370_010 |
0.0919131637 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 7 |
Hb_000910_030 |
0.0983378141 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000103_300 |
0.099298063 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 9 |
Hb_003607_090 |
0.0994896511 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 10 |
Hb_000152_570 |
0.1009503451 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 11 |
Hb_000419_140 |
0.1046586801 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 12 |
Hb_004531_160 |
0.1053482065 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_009456_010 |
0.1058806482 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X3 [Populus euphratica] |
| 14 |
Hb_002566_020 |
0.1105816784 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 15 |
Hb_113766_010 |
0.1114326451 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |
| 16 |
Hb_006154_010 |
0.1137978546 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000193_070 |
0.1157943477 |
- |
- |
hypothetical protein JCGZ_25373 [Jatropha curcas] |
| 18 |
Hb_031224_010 |
0.1173517227 |
- |
- |
- |
| 19 |
Hb_000076_280 |
0.1180831884 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 20 |
Hb_003633_170 |
0.1182470696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |