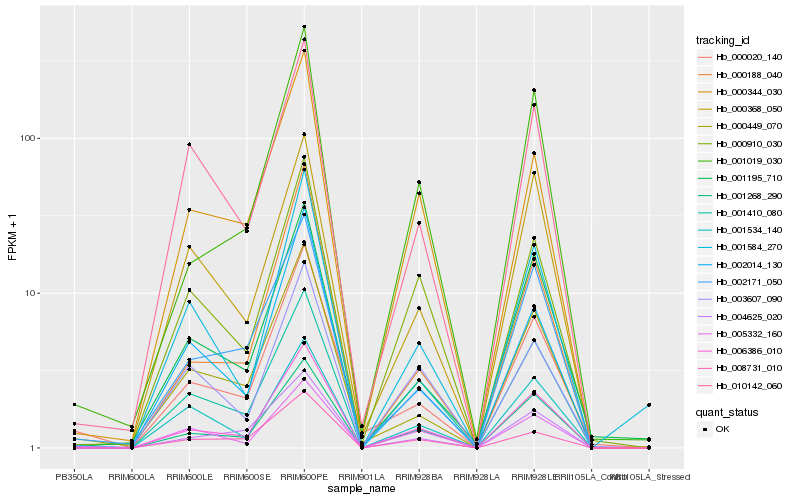
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006386_010 |
0.0 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 2 |
Hb_001268_290 |
0.047446408 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 3 |
Hb_001019_030 |
0.0912816912 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 4 |
Hb_001410_080 |
0.0930436811 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_000910_030 |
0.0953653733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000344_030 |
0.0982852118 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 7 |
Hb_001195_710 |
0.1025370862 |
- |
- |
glycosyltransferase family GT8 protein [Populus deltoides] |
| 8 |
Hb_001584_270 |
0.1094679287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001534_140 |
0.1104141653 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 10 |
Hb_000188_040 |
0.11166598 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 11 |
Hb_002014_130 |
0.1132889887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002171_050 |
0.1133170619 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 13 |
Hb_005332_160 |
0.1136370015 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 14 |
Hb_000449_070 |
0.1149820146 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000020_140 |
0.1150326498 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 16 |
Hb_010142_060 |
0.1152485987 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 17 |
Hb_004625_020 |
0.1178729078 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 18 |
Hb_003607_090 |
0.1203383583 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 19 |
Hb_008731_010 |
0.1208781822 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 20 |
Hb_000368_050 |
0.1219269308 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |