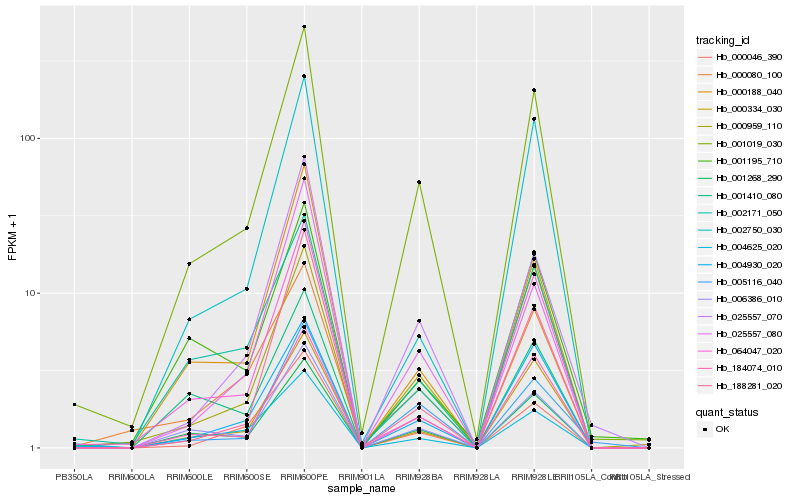
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001019_030 |
0.0 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 2 |
Hb_006386_010 |
0.0912816912 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 3 |
Hb_001268_290 |
0.0923136851 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 4 |
Hb_184074_010 |
0.0949372593 |
- |
- |
PREDICTED: uncharacterized protein LOC105639054 [Jatropha curcas] |
| 5 |
Hb_000334_030 |
0.1029563322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004930_020 |
0.1046388138 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 7 |
Hb_005116_040 |
0.1133626396 |
- |
- |
PREDICTED: uncharacterized protein LOC105131445 [Populus euphratica] |
| 8 |
Hb_025557_070 |
0.1161466577 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |
| 9 |
Hb_000046_390 |
0.1169654593 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 10 |
Hb_064047_020 |
0.1178778017 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 11 |
Hb_000080_100 |
0.1233662631 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 12 |
Hb_000188_040 |
0.1253089707 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 13 |
Hb_000959_110 |
0.1281077127 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 14 |
Hb_188281_020 |
0.1304088624 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 15 |
Hb_025557_080 |
0.1328091556 |
transcription factor |
TF Family: zf-HD |
- |
| 16 |
Hb_002171_050 |
0.1330354962 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 17 |
Hb_001195_710 |
0.137285877 |
- |
- |
glycosyltransferase family GT8 protein [Populus deltoides] |
| 18 |
Hb_004625_020 |
0.1387924278 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 19 |
Hb_001410_080 |
0.1394056661 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_002750_030 |
0.1410294451 |
- |
- |
PREDICTED: protein IRX15-LIKE-like [Populus euphratica] |