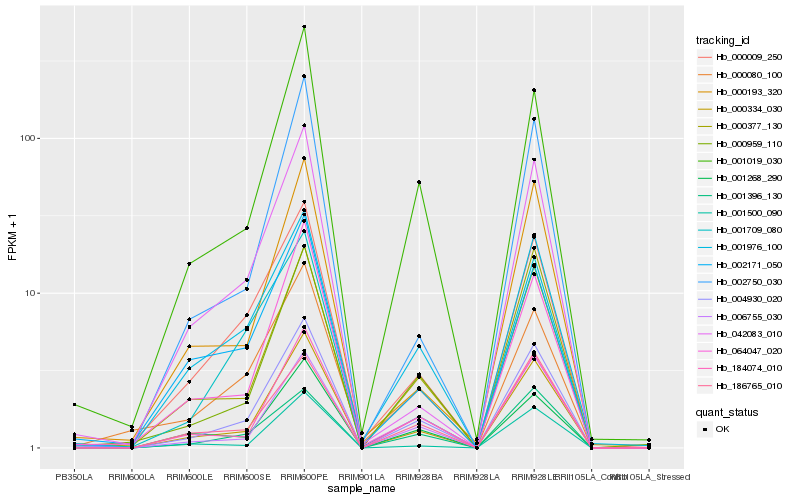
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004930_020 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 2 |
Hb_000334_030 |
0.0584374377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000959_110 |
0.0757933802 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 4 |
Hb_184074_010 |
0.0874971914 |
- |
- |
PREDICTED: uncharacterized protein LOC105639054 [Jatropha curcas] |
| 5 |
Hb_001976_100 |
0.0916900339 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000009_250 |
0.1035153711 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 7 |
Hb_001019_030 |
0.1046388138 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 8 |
Hb_006755_030 |
0.1050342588 |
- |
- |
BURP domain-containing protein, putative [Theobroma cacao] |
| 9 |
Hb_000377_130 |
0.1077714075 |
- |
- |
hypothetical protein POPTR_0016s00790g [Populus trichocarpa] |
| 10 |
Hb_001396_130 |
0.1189455333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000080_100 |
0.1190439587 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 12 |
Hb_002750_030 |
0.1215119981 |
- |
- |
PREDICTED: protein IRX15-LIKE-like [Populus euphratica] |
| 13 |
Hb_000193_320 |
0.1229772924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_186765_010 |
0.1238654326 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 15 |
Hb_001268_290 |
0.124573535 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 16 |
Hb_002171_050 |
0.1256625641 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 17 |
Hb_001500_090 |
0.1282873508 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001709_080 |
0.1282896375 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_064047_020 |
0.1284101687 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0003s13190g [Populus trichocarpa] |
| 20 |
Hb_042083_010 |
0.1314198426 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Jatropha curcas] |