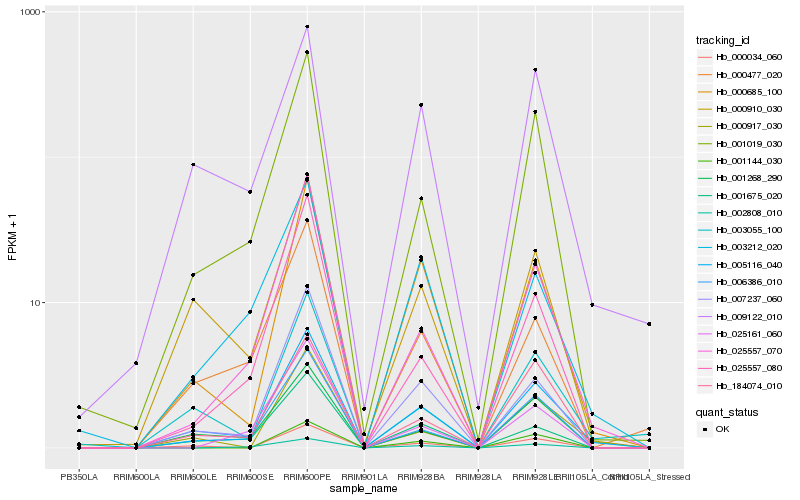
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005116_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105131445 [Populus euphratica] |
| 2 |
Hb_000685_100 |
0.1104090826 |
- |
- |
hypothetical protein POPTR_0001s10630g [Populus trichocarpa] |
| 3 |
Hb_001019_030 |
0.1133626396 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 4 |
Hb_025557_070 |
0.1159529742 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |
| 5 |
Hb_007237_060 |
0.1254412065 |
- |
- |
- |
| 6 |
Hb_000917_030 |
0.1386215987 |
- |
- |
PREDICTED: uncharacterized protein LOC102629571 [Citrus sinensis] |
| 7 |
Hb_000034_060 |
0.1454152387 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 8 |
Hb_006386_010 |
0.1456573684 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 9 |
Hb_001268_290 |
0.1500621524 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 10 |
Hb_001675_020 |
0.1510063746 |
- |
- |
- |
| 11 |
Hb_184074_010 |
0.1512368634 |
- |
- |
PREDICTED: uncharacterized protein LOC105639054 [Jatropha curcas] |
| 12 |
Hb_000910_030 |
0.1538357669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002808_010 |
0.1541315487 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 14 |
Hb_025557_080 |
0.1544442634 |
transcription factor |
TF Family: zf-HD |
- |
| 15 |
Hb_003212_020 |
0.1589191259 |
- |
- |
hypothetical protein PHAVU_009G114800g [Phaseolus vulgaris] |
| 16 |
Hb_001144_030 |
0.1589551581 |
- |
- |
hypothetical protein CICLE_v10023280mg, partial [Citrus clementina] |
| 17 |
Hb_003055_100 |
0.1600024142 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 18 |
Hb_000477_020 |
0.1613422631 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |
| 19 |
Hb_025161_060 |
0.1618653494 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 20 |
Hb_009122_010 |
0.162717085 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |