| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
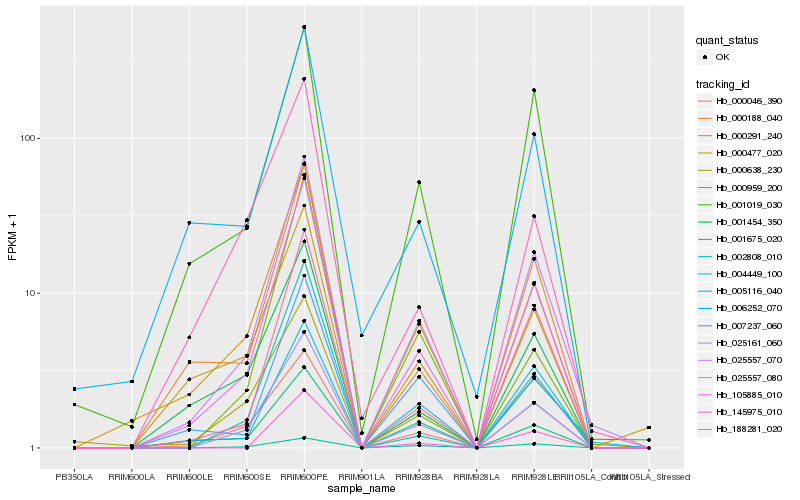
Hb_025161_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 2 |
Hb_025557_080 |
0.0798798178 |
transcription factor |
TF Family: zf-HD |
- |
| 3 |
Hb_025557_070 |
0.0826604548 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |
| 4 |
Hb_000046_390 |
0.0953366402 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 5 |
Hb_000638_230 |
0.1182889623 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 6 |
Hb_000291_240 |
0.1284382473 |
- |
- |
hypothetical protein POPTR_0005s16430g [Populus trichocarpa] |
| 7 |
Hb_002808_010 |
0.1427263259 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 8 |
Hb_004449_100 |
0.1441567085 |
- |
- |
- |
| 9 |
Hb_188281_020 |
0.1456570985 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 10 |
Hb_001019_030 |
0.1484172286 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 11 |
Hb_001454_350 |
0.1531026044 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 12 |
Hb_007237_060 |
0.1532646162 |
- |
- |
- |
| 13 |
Hb_000188_040 |
0.1549581886 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 14 |
Hb_000477_020 |
0.1562192877 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |
| 15 |
Hb_145975_010 |
0.1596615218 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_005116_040 |
0.1618653494 |
- |
- |
PREDICTED: uncharacterized protein LOC105131445 [Populus euphratica] |
| 17 |
Hb_001675_020 |
0.1660999489 |
- |
- |
- |
| 18 |
Hb_105885_010 |
0.1664598991 |
- |
- |
PREDICTED: uncharacterized protein LOC103441690 [Malus domestica] |
| 19 |
Hb_000959_200 |
0.1668224558 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 20 |
Hb_006252_070 |
0.1685172259 |
- |
- |
hypothetical protein JCGZ_17254 [Jatropha curcas] |