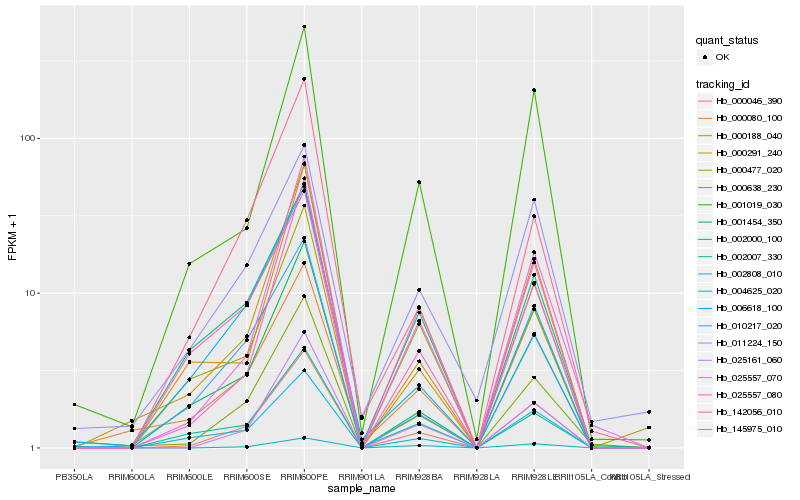
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_390 |
0.0 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 2 |
Hb_025161_060 |
0.0953366402 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 3 |
Hb_000638_230 |
0.0998243464 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 4 |
Hb_001454_350 |
0.1113283463 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 5 |
Hb_025557_070 |
0.111580809 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |
| 6 |
Hb_025557_080 |
0.1164539813 |
transcription factor |
TF Family: zf-HD |
- |
| 7 |
Hb_001019_030 |
0.1169654593 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 8 |
Hb_142056_010 |
0.1277236691 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002000_100 |
0.1285654068 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 10 |
Hb_004625_020 |
0.130581665 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 11 |
Hb_000080_100 |
0.1315307623 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 12 |
Hb_000291_240 |
0.1333338262 |
- |
- |
hypothetical protein POPTR_0005s16430g [Populus trichocarpa] |
| 13 |
Hb_010217_020 |
0.1352656463 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 14 |
Hb_011224_150 |
0.1407847301 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002808_010 |
0.1410548789 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 16 |
Hb_000188_040 |
0.1416256977 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 17 |
Hb_002007_330 |
0.1434300339 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 18 |
Hb_145975_010 |
0.1475195779 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_006618_100 |
0.1504802971 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 20 |
Hb_000477_020 |
0.1505027605 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |