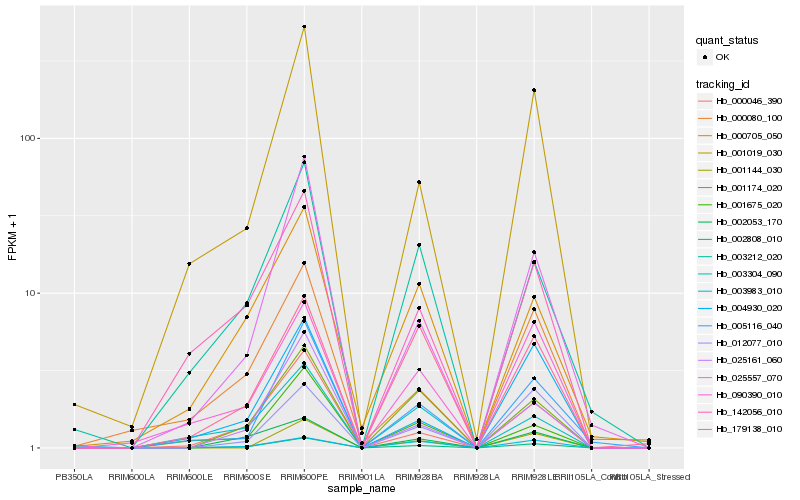
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002808_010 |
0.0 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 2 |
Hb_001174_020 |
0.1014634529 |
- |
- |
- |
| 3 |
Hb_002053_170 |
0.1361495695 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_000046_390 |
0.1410548789 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 5 |
Hb_025161_060 |
0.1427263259 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 6 |
Hb_005116_040 |
0.1541315487 |
- |
- |
PREDICTED: uncharacterized protein LOC105131445 [Populus euphratica] |
| 7 |
Hb_001019_030 |
0.1561282551 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 8 |
Hb_003212_020 |
0.1584107151 |
- |
- |
hypothetical protein PHAVU_009G114800g [Phaseolus vulgaris] |
| 9 |
Hb_000705_050 |
0.1595821401 |
- |
- |
PREDICTED: uncharacterized protein C167.05 [Vitis vinifera] |
| 10 |
Hb_142056_010 |
0.1624930017 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_090390_010 |
0.1645801302 |
- |
- |
hypothetical protein POPTR_0006s28850g [Populus trichocarpa] |
| 12 |
Hb_012077_010 |
0.1678593792 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 13 |
Hb_025557_070 |
0.1732861788 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |
| 14 |
Hb_179138_010 |
0.1735282595 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 15 |
Hb_000080_100 |
0.1779197975 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 16 |
Hb_001144_030 |
0.1793153806 |
- |
- |
hypothetical protein CICLE_v10023280mg, partial [Citrus clementina] |
| 17 |
Hb_001675_020 |
0.180221806 |
- |
- |
- |
| 18 |
Hb_003304_090 |
0.1809304442 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 19 |
Hb_004930_020 |
0.1810312659 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 20 |
Hb_003983_010 |
0.1831592322 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |