| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
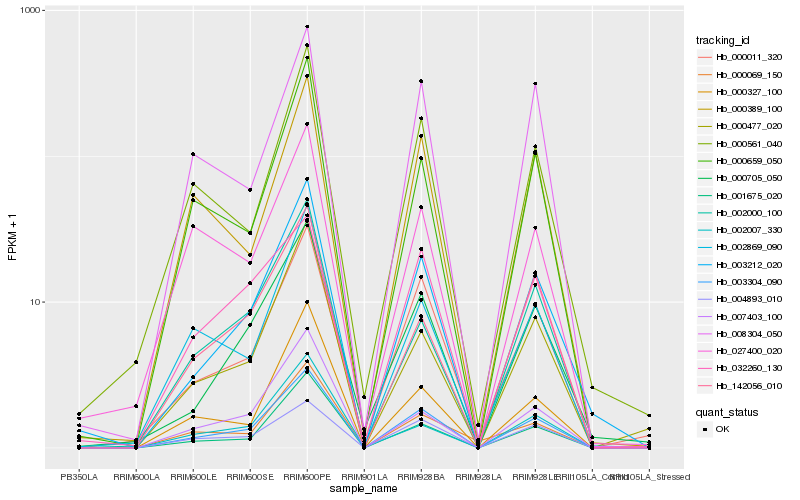
Hb_003304_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 2 |
Hb_000011_320 |
0.0917500725 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 3 |
Hb_003212_020 |
0.0922248014 |
- |
- |
hypothetical protein PHAVU_009G114800g [Phaseolus vulgaris] |
| 4 |
Hb_004893_010 |
0.1005710125 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 5 |
Hb_001675_020 |
0.104443778 |
- |
- |
- |
| 6 |
Hb_000705_050 |
0.1111280661 |
- |
- |
PREDICTED: uncharacterized protein C167.05 [Vitis vinifera] |
| 7 |
Hb_000659_050 |
0.1163849239 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 8 |
Hb_008304_050 |
0.1207882484 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 9 |
Hb_000389_100 |
0.1212023572 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 10 |
Hb_000561_040 |
0.1220217493 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 11 |
Hb_002869_090 |
0.1260800261 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 12 |
Hb_007403_100 |
0.1291174539 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 13 |
Hb_142056_010 |
0.131694459 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_032260_130 |
0.1358190474 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 15 |
Hb_000477_020 |
0.1371537093 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |
| 16 |
Hb_002007_330 |
0.1386586796 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 17 |
Hb_000069_150 |
0.1390629479 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 18 |
Hb_002000_100 |
0.1391118723 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 19 |
Hb_000327_100 |
0.1397092301 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 20 |
Hb_027400_020 |
0.1403553756 |
- |
- |
PREDICTED: uncharacterized protein At5g22580 isoform X2 [Jatropha curcas] |