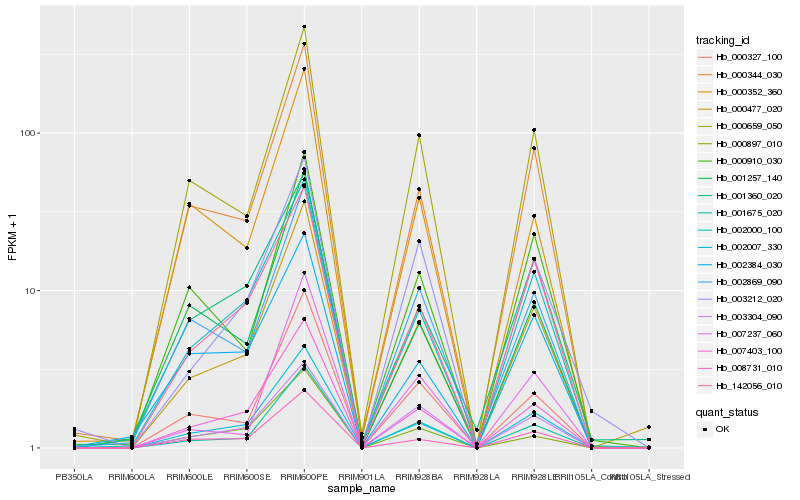
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000477_020 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |
| 2 |
Hb_001675_020 |
0.0765772611 |
- |
- |
- |
| 3 |
Hb_007403_100 |
0.0818421464 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 4 |
Hb_002007_330 |
0.0849421389 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 5 |
Hb_000344_030 |
0.0901520298 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 6 |
Hb_000327_100 |
0.0970267355 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 7 |
Hb_002000_100 |
0.1011307141 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 8 |
Hb_000659_050 |
0.101694677 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 9 |
Hb_008731_010 |
0.1054178083 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 10 |
Hb_002869_090 |
0.1169562591 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 11 |
Hb_142056_010 |
0.1203233789 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007237_060 |
0.1272351151 |
- |
- |
- |
| 13 |
Hb_001360_020 |
0.1336282136 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 14 |
Hb_003212_020 |
0.1345205037 |
- |
- |
hypothetical protein PHAVU_009G114800g [Phaseolus vulgaris] |
| 15 |
Hb_003304_090 |
0.1371537093 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 16 |
Hb_000910_030 |
0.1379395743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001257_140 |
0.1386021397 |
- |
- |
Gb:AAF02129.1, putative [Theobroma cacao] |
| 18 |
Hb_000352_360 |
0.13885542 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 19 |
Hb_000897_010 |
0.1421415786 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 20 |
Hb_002384_030 |
0.1430224443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |