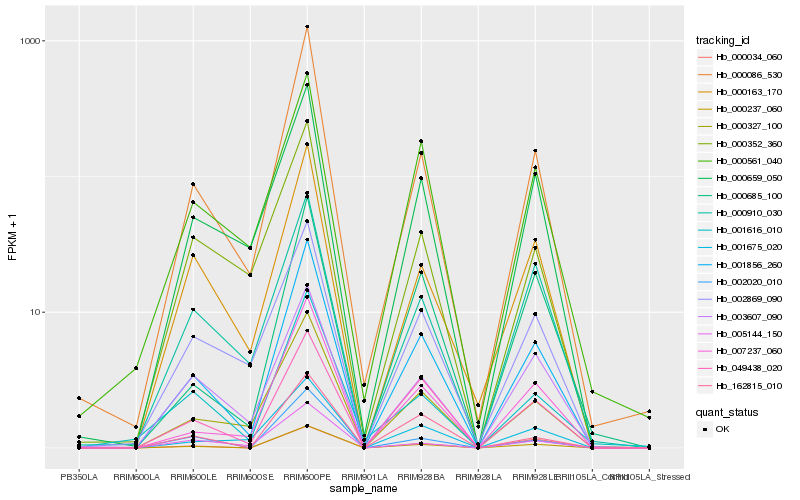
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001856_260 |
0.0 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 2 |
Hb_000237_060 |
0.062279553 |
- |
- |
hypothetical protein PRUPE_ppa022052mg, partial [Prunus persica] |
| 3 |
Hb_000086_530 |
0.0761862315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007237_060 |
0.088866475 |
- |
- |
- |
| 5 |
Hb_000327_100 |
0.096146754 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 6 |
Hb_049438_020 |
0.1113988938 |
- |
- |
PREDICTED: peroxidase 20 [Jatropha curcas] |
| 7 |
Hb_000163_170 |
0.1187834472 |
- |
- |
- |
| 8 |
Hb_000659_050 |
0.1211243052 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 9 |
Hb_001675_020 |
0.1243987485 |
- |
- |
- |
| 10 |
Hb_002020_010 |
0.1271547846 |
- |
- |
kinase-like protein [Corylus avellana] |
| 11 |
Hb_000685_100 |
0.1285429935 |
- |
- |
hypothetical protein POPTR_0001s10630g [Populus trichocarpa] |
| 12 |
Hb_162815_010 |
0.1299372307 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 13 |
Hb_002869_090 |
0.1313467921 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 14 |
Hb_003607_090 |
0.1382876784 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 15 |
Hb_000910_030 |
0.1383037374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001616_010 |
0.1403678971 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 17 |
Hb_000561_040 |
0.1413509628 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 18 |
Hb_000034_060 |
0.1414611536 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 19 |
Hb_005144_150 |
0.148140869 |
- |
- |
hypothetical protein VITISV_013572 [Vitis vinifera] |
| 20 |
Hb_000352_360 |
0.1484013488 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |