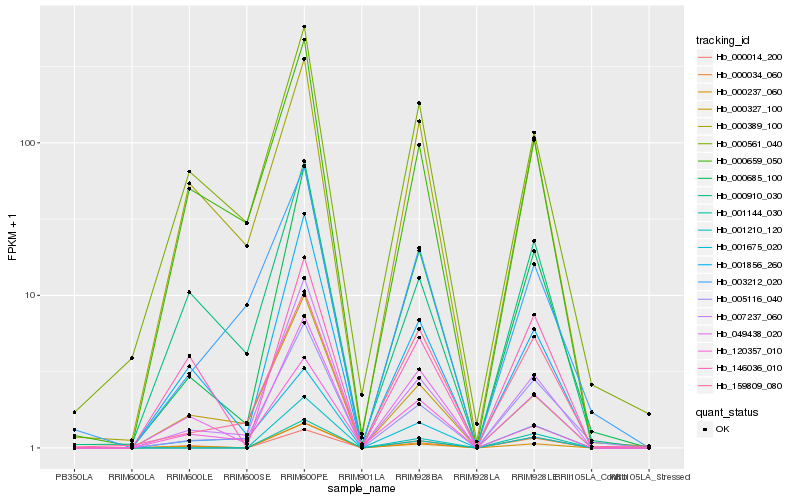
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000685_100 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s10630g [Populus trichocarpa] |
| 2 |
Hb_007237_060 |
0.1091150561 |
- |
- |
- |
| 3 |
Hb_005116_040 |
0.1104090826 |
- |
- |
PREDICTED: uncharacterized protein LOC105131445 [Populus euphratica] |
| 4 |
Hb_049438_020 |
0.1231581075 |
- |
- |
PREDICTED: peroxidase 20 [Jatropha curcas] |
| 5 |
Hb_000034_060 |
0.1252046189 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 6 |
Hb_001856_260 |
0.1285429935 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 7 |
Hb_000237_060 |
0.1479318091 |
- |
- |
hypothetical protein PRUPE_ppa022052mg, partial [Prunus persica] |
| 8 |
Hb_001675_020 |
0.148841791 |
- |
- |
- |
| 9 |
Hb_001144_030 |
0.1535786596 |
- |
- |
hypothetical protein CICLE_v10023280mg, partial [Citrus clementina] |
| 10 |
Hb_159809_080 |
0.1600799873 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 11 |
Hb_001210_120 |
0.1604012772 |
- |
- |
hypothetical protein JCGZ_06226 [Jatropha curcas] |
| 12 |
Hb_000561_040 |
0.1606445394 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 13 |
Hb_000659_050 |
0.1644873983 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 14 |
Hb_000014_200 |
0.1667245479 |
- |
- |
PREDICTED: uncharacterized protein LOC105775355 [Gossypium raimondii] |
| 15 |
Hb_000327_100 |
0.1671674312 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 16 |
Hb_003212_020 |
0.1689071759 |
- |
- |
hypothetical protein PHAVU_009G114800g [Phaseolus vulgaris] |
| 17 |
Hb_000910_030 |
0.171857749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_120357_010 |
0.1727107503 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000389_100 |
0.1731034264 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 20 |
Hb_146036_010 |
0.1756693626 |
- |
- |
hypothetical protein B456_005G052500 [Gossypium raimondii] |