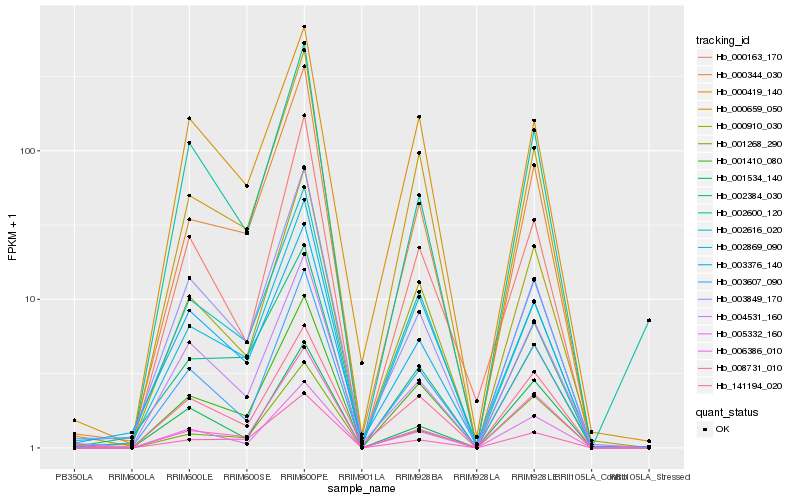
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000910_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003607_090 |
0.0376402668 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 3 |
Hb_005332_160 |
0.0676821584 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 4 |
Hb_000659_050 |
0.0729677806 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 5 |
Hb_001410_080 |
0.0744516808 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_000344_030 |
0.0838774406 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 7 |
Hb_002869_090 |
0.0910636673 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 8 |
Hb_006386_010 |
0.0953653733 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 9 |
Hb_000163_170 |
0.0960712542 |
- |
- |
- |
| 10 |
Hb_141194_020 |
0.0983378141 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 11 |
Hb_004531_160 |
0.1019936254 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001268_290 |
0.1090164906 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 13 |
Hb_003849_170 |
0.1156768345 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 14 |
Hb_003376_140 |
0.1158031211 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 15 |
Hb_001534_140 |
0.1179521606 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 16 |
Hb_002616_020 |
0.1179852028 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 17 |
Hb_002600_120 |
0.1194911507 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 18 |
Hb_000419_140 |
0.1206829277 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 19 |
Hb_008731_010 |
0.122558404 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 20 |
Hb_002384_030 |
0.1269421748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |