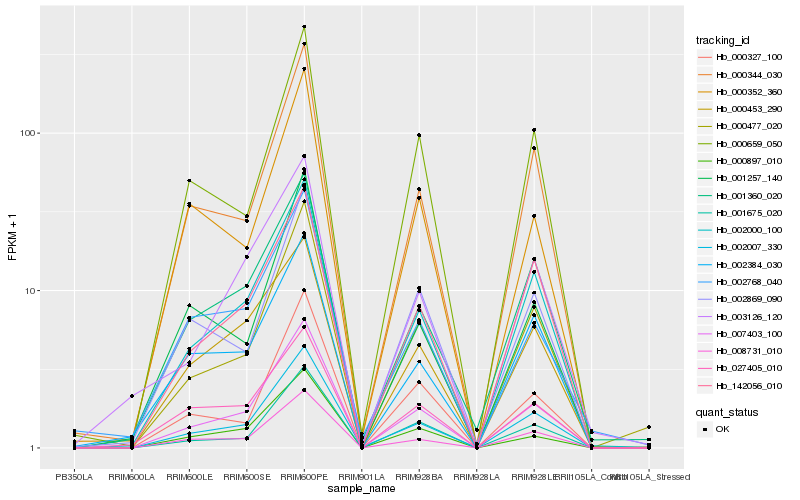
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007403_100 |
0.0 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 2 |
Hb_002007_330 |
0.0469282002 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 3 |
Hb_002000_100 |
0.0671847011 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 4 |
Hb_008731_010 |
0.072289679 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 5 |
Hb_000477_020 |
0.0818421464 |
- |
- |
hypothetical protein EUGRSUZ_C011052, partial [Eucalyptus grandis] |
| 6 |
Hb_000897_010 |
0.0828690629 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 7 |
Hb_000344_030 |
0.0867687264 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 8 |
Hb_001675_020 |
0.0894055555 |
- |
- |
- |
| 9 |
Hb_000327_100 |
0.1023540203 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 10 |
Hb_142056_010 |
0.1038383868 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000453_290 |
0.108818859 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 12 |
Hb_000659_050 |
0.1099536349 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 13 |
Hb_001360_020 |
0.1107615395 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 14 |
Hb_002869_090 |
0.1138484409 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 15 |
Hb_000352_360 |
0.1158243625 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 16 |
Hb_027405_010 |
0.1160373088 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 17 |
Hb_002384_030 |
0.1166905387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001257_140 |
0.1195871032 |
- |
- |
Gb:AAF02129.1, putative [Theobroma cacao] |
| 19 |
Hb_003126_120 |
0.1235274792 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 20 |
Hb_002768_040 |
0.123905065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |