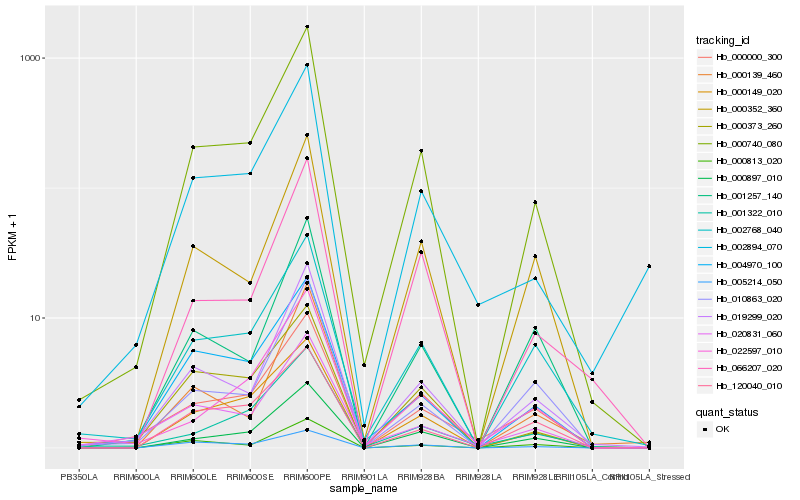
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000740_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_019299_020 |
0.0903006926 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 3 |
Hb_000897_010 |
0.0957290062 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 4 |
Hb_000149_020 |
0.1019935483 |
- |
- |
Proline-rich receptor-like protein kinase PERK14 [Glycine soja] |
| 5 |
Hb_002768_040 |
0.109889563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004970_100 |
0.1163169305 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 7 |
Hb_000352_360 |
0.1193356019 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 8 |
Hb_001322_010 |
0.1246321029 |
- |
- |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 9 |
Hb_000139_460 |
0.1253230867 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 10 |
Hb_066207_020 |
0.1272380714 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 11 |
Hb_010863_020 |
0.1293183999 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000000_300 |
0.1314131122 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 13 |
Hb_000373_260 |
0.1315209922 |
- |
- |
hypothetical protein POPTR_0002s06820g [Populus trichocarpa] |
| 14 |
Hb_002894_070 |
0.1326987798 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein B [Jatropha curcas] |
| 15 |
Hb_000813_020 |
0.1346906168 |
- |
- |
PREDICTED: metacaspase-3-like [Jatropha curcas] |
| 16 |
Hb_022597_010 |
0.1353854311 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 17 |
Hb_005214_050 |
0.1368110966 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 18 |
Hb_120040_010 |
0.1369330747 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 19 |
Hb_001257_140 |
0.1371795218 |
- |
- |
Gb:AAF02129.1, putative [Theobroma cacao] |
| 20 |
Hb_020831_060 |
0.1373500466 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |