| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
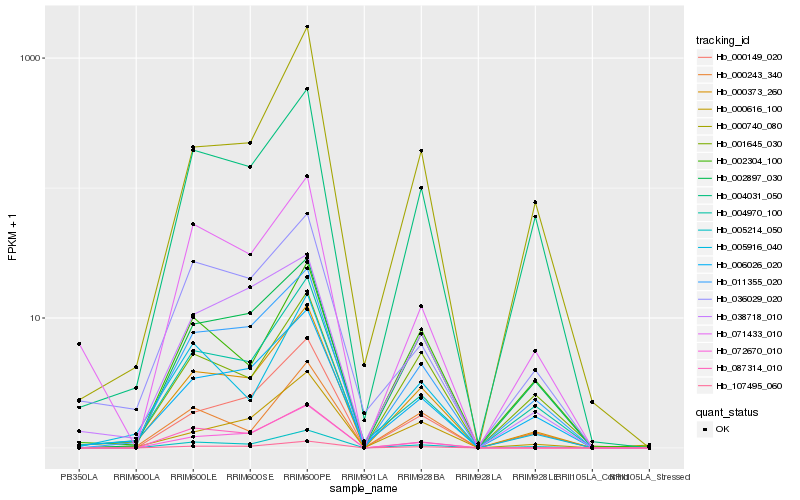
Hb_000373_260 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s06820g [Populus trichocarpa] |
| 2 |
Hb_005214_050 |
0.0906182332 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 3 |
Hb_011355_020 |
0.0926204514 |
- |
- |
extensin, proline-rich protein, putative [Ricinus communis] |
| 4 |
Hb_006026_020 |
0.0938422679 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g48800 [Jatropha curcas] |
| 5 |
Hb_004970_100 |
0.1058049534 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 6 |
Hb_004031_050 |
0.1067800348 |
- |
- |
sinapyl alcohol dehydrogenase-like protein [Populus tremula x Populus tremuloides] |
| 7 |
Hb_000149_020 |
0.1139340486 |
- |
- |
Proline-rich receptor-like protein kinase PERK14 [Glycine soja] |
| 8 |
Hb_107495_060 |
0.1154013063 |
- |
- |
Cell division protein kinase, putative [Ricinus communis] |
| 9 |
Hb_002897_030 |
0.1160477539 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 10 |
Hb_000740_080 |
0.1315209922 |
- |
- |
- |
| 11 |
Hb_001645_030 |
0.1326372244 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1b [Jatropha curcas] |
| 12 |
Hb_005916_040 |
0.1351420172 |
- |
- |
PREDICTED: auxin transporter-like protein 5 [Populus euphratica] |
| 13 |
Hb_000616_100 |
0.1390365407 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000243_340 |
0.1424834825 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 15 |
Hb_072670_010 |
0.1428177073 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 16 |
Hb_002304_100 |
0.1464145069 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 17 |
Hb_087314_010 |
0.1483735351 |
- |
- |
- |
| 18 |
Hb_036029_020 |
0.1485337463 |
- |
- |
PREDICTED: patellin-3-like [Jatropha curcas] |
| 19 |
Hb_071433_010 |
0.1505865018 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 20 |
Hb_038718_010 |
0.1515409736 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |