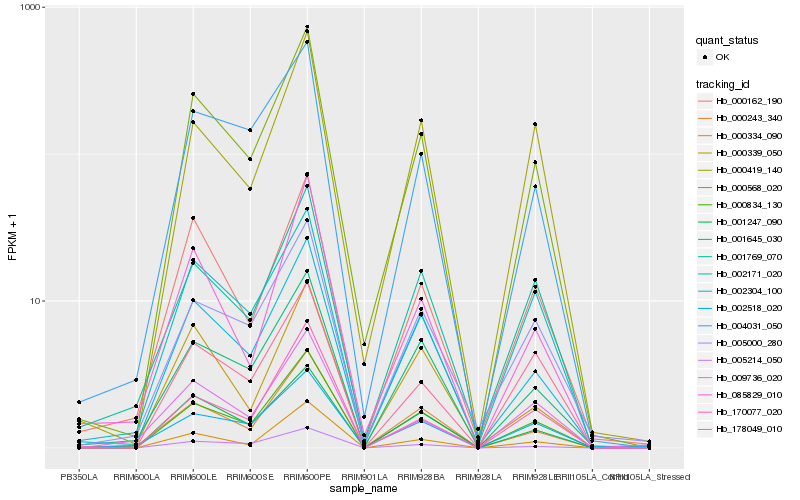
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000568_020 |
0.0 |
- |
- |
PREDICTED: probable aquaporin PIP2-5 [Jatropha curcas] |
| 2 |
Hb_000834_130 |
0.0646876051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002304_100 |
0.0764138758 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 4 |
Hb_000243_340 |
0.0846439397 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 5 |
Hb_001247_090 |
0.0879373077 |
- |
- |
PREDICTED: uncharacterized protein LOC105647395 [Jatropha curcas] |
| 6 |
Hb_005214_050 |
0.0980275508 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 7 |
Hb_004031_050 |
0.099183543 |
- |
- |
sinapyl alcohol dehydrogenase-like protein [Populus tremula x Populus tremuloides] |
| 8 |
Hb_001645_030 |
0.1028206326 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1b [Jatropha curcas] |
| 9 |
Hb_000162_190 |
0.1047810395 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 10 |
Hb_002518_020 |
0.1110862979 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 11 |
Hb_002171_020 |
0.1113587453 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 12 |
Hb_000334_090 |
0.1128946067 |
- |
- |
hypothetical protein JCGZ_16789 [Jatropha curcas] |
| 13 |
Hb_005000_280 |
0.1167503935 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 14 |
Hb_001769_070 |
0.1210006998 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_170077_020 |
0.1219386321 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 16 |
Hb_000419_140 |
0.1231787541 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 17 |
Hb_178049_010 |
0.1238327813 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_085829_010 |
0.1253263146 |
- |
- |
PREDICTED: homeobox protein 2-like [Jatropha curcas] |
| 19 |
Hb_000339_050 |
0.1257784897 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 20 |
Hb_009736_020 |
0.1277317557 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG13 homolog [Jatropha curcas] |