| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
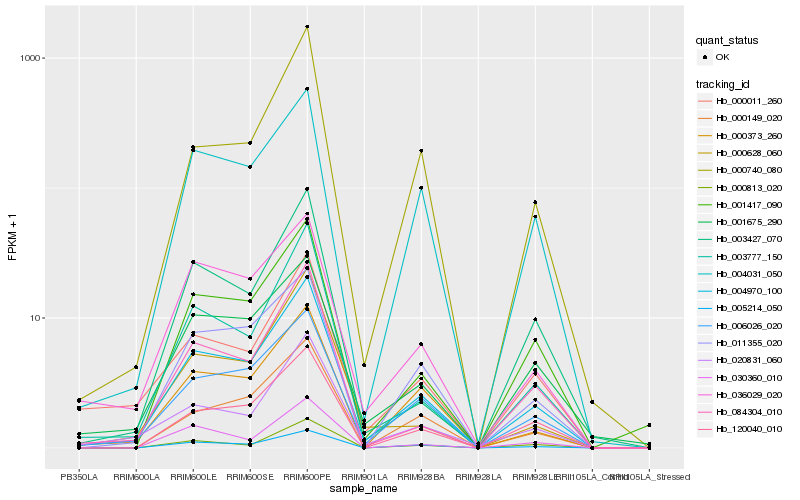
Hb_004970_100 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-4 [Jatropha curcas] |
| 2 |
Hb_006026_020 |
0.0856092234 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g48800 [Jatropha curcas] |
| 3 |
Hb_003777_150 |
0.1006178887 |
- |
- |
PREDICTED: uncharacterized protein LOC105640938 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005214_050 |
0.1019947108 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 5 |
Hb_000373_260 |
0.1058049534 |
- |
- |
hypothetical protein POPTR_0002s06820g [Populus trichocarpa] |
| 6 |
Hb_001417_090 |
0.1066211177 |
- |
- |
PREDICTED: uncharacterized protein LOC105650625 [Jatropha curcas] |
| 7 |
Hb_004031_050 |
0.1106046397 |
- |
- |
sinapyl alcohol dehydrogenase-like protein [Populus tremula x Populus tremuloides] |
| 8 |
Hb_084304_010 |
0.1113276809 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X2 [Jatropha curcas] |
| 9 |
Hb_120040_010 |
0.1124344529 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_020831_060 |
0.1132573792 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 11 |
Hb_011355_020 |
0.1161399985 |
- |
- |
extensin, proline-rich protein, putative [Ricinus communis] |
| 12 |
Hb_000740_080 |
0.1163169305 |
- |
- |
- |
| 13 |
Hb_001675_290 |
0.1215386016 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000011_260 |
0.1215733305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000628_060 |
0.1218221952 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 16 |
Hb_000149_020 |
0.1241357456 |
- |
- |
Proline-rich receptor-like protein kinase PERK14 [Glycine soja] |
| 17 |
Hb_036029_020 |
0.1251644036 |
- |
- |
PREDICTED: patellin-3-like [Jatropha curcas] |
| 18 |
Hb_003427_070 |
0.1303412328 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 19 |
Hb_030360_010 |
0.1332620914 |
- |
- |
- |
| 20 |
Hb_000813_020 |
0.1349928677 |
- |
- |
PREDICTED: metacaspase-3-like [Jatropha curcas] |