| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
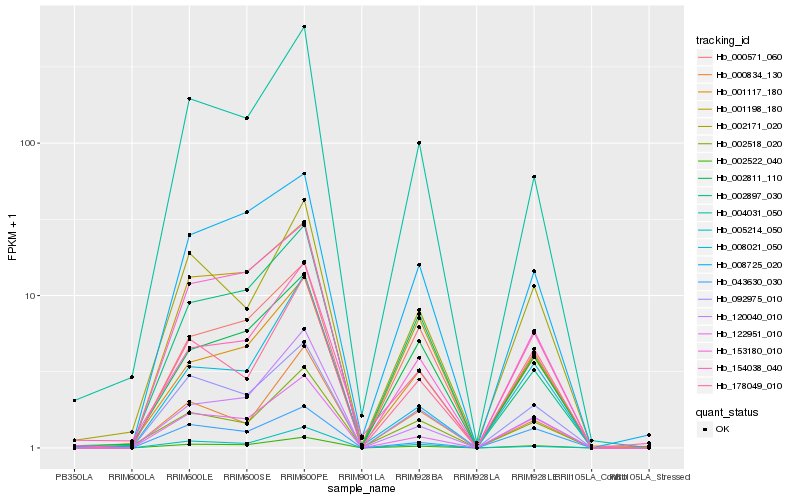
Hb_002522_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 2 |
Hb_092975_010 |
0.0724770839 |
- |
- |
hypothetical protein JCGZ_04359 [Jatropha curcas] |
| 3 |
Hb_122951_010 |
0.0745129546 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 4 |
Hb_004031_050 |
0.0889609616 |
- |
- |
sinapyl alcohol dehydrogenase-like protein [Populus tremula x Populus tremuloides] |
| 5 |
Hb_001117_180 |
0.0945750546 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 6 |
Hb_153180_010 |
0.0973256823 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 7 |
Hb_178049_010 |
0.1035201218 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_120040_010 |
0.1052461532 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_002171_020 |
0.109784666 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 10 |
Hb_001198_180 |
0.1135187491 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 11 |
Hb_008725_020 |
0.1155927713 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 12 |
Hb_000571_060 |
0.1162772277 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 13 |
Hb_002518_020 |
0.1167172755 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 14 |
Hb_008021_050 |
0.1174212973 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_043630_030 |
0.1188371035 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 16 |
Hb_002811_110 |
0.1189830003 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 17 |
Hb_000834_130 |
0.1234592937 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005214_050 |
0.1240871129 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 19 |
Hb_002897_030 |
0.1242156869 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 20 |
Hb_154038_040 |
0.1244572738 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |