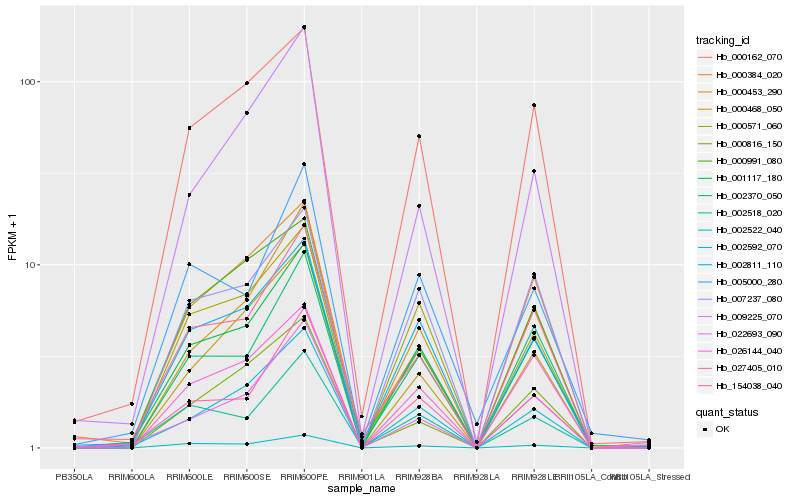
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001117_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 2 |
Hb_154038_040 |
0.0751303064 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 3 |
Hb_002811_110 |
0.0828898661 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 4 |
Hb_027405_010 |
0.0932313251 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 5 |
Hb_000162_070 |
0.0937709907 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 6 |
Hb_002592_070 |
0.0939027557 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 7 |
Hb_002522_040 |
0.0945750546 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 8 |
Hb_000453_290 |
0.0969597366 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 9 |
Hb_002518_020 |
0.1025891138 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 10 |
Hb_000384_020 |
0.105337997 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |
| 11 |
Hb_026144_040 |
0.1069148997 |
- |
- |
- |
| 12 |
Hb_002370_050 |
0.1077024982 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 13 |
Hb_022693_090 |
0.1077910276 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000468_050 |
0.1083634775 |
- |
- |
- |
| 15 |
Hb_000571_060 |
0.1116636338 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 16 |
Hb_007237_080 |
0.1123498306 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_005000_280 |
0.1140129142 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 18 |
Hb_009225_070 |
0.1156604628 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 19 |
Hb_000816_150 |
0.1190959823 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 20 |
Hb_000991_080 |
0.1193267417 |
- |
- |
kinase, putative [Ricinus communis] |