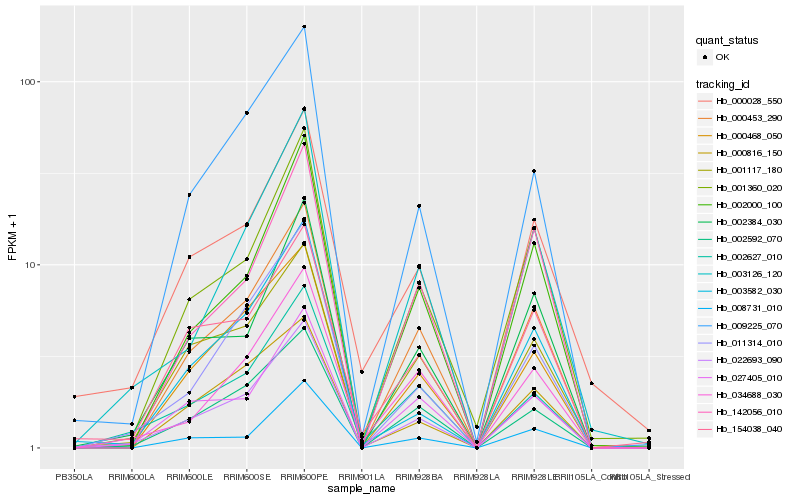
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022693_090 |
0.0 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000453_290 |
0.0846345459 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 3 |
Hb_002384_030 |
0.0886726827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009225_070 |
0.0947288395 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 5 |
Hb_001360_020 |
0.0970888074 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 6 |
Hb_011314_010 |
0.0982784085 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002592_070 |
0.0995139018 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 8 |
Hb_003126_120 |
0.1015204821 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 9 |
Hb_027405_010 |
0.1016043001 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 10 |
Hb_002000_100 |
0.1042760094 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 11 |
Hb_001117_180 |
0.1077910276 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000468_050 |
0.1117257617 |
- |
- |
- |
| 13 |
Hb_002627_010 |
0.1131458711 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 14 |
Hb_003582_030 |
0.1136608439 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 15 |
Hb_154038_040 |
0.1147180847 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 16 |
Hb_034688_030 |
0.120924555 |
- |
- |
PREDICTED: uncharacterized protein LOC105642049 [Jatropha curcas] |
| 17 |
Hb_000028_550 |
0.1224010689 |
- |
- |
unknown [Medicago truncatula] |
| 18 |
Hb_008731_010 |
0.1234873968 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |
| 19 |
Hb_142056_010 |
0.1235110259 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000816_150 |
0.1239137823 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |