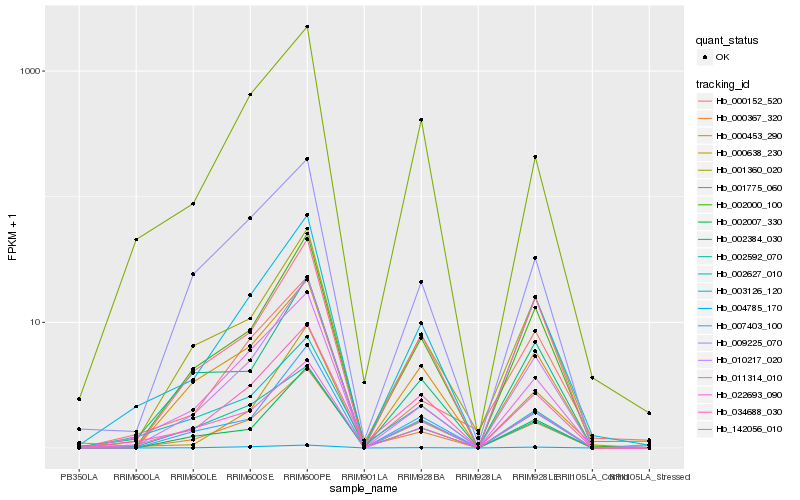
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003126_120 |
0.0 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 2 |
Hb_034688_030 |
0.0839442969 |
- |
- |
PREDICTED: uncharacterized protein LOC105642049 [Jatropha curcas] |
| 3 |
Hb_002000_100 |
0.0995718184 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 4 |
Hb_022693_090 |
0.1015204821 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_011314_010 |
0.1073789758 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001360_020 |
0.1191053041 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 7 |
Hb_000367_320 |
0.1214625013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007403_100 |
0.1235274792 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 9 |
Hb_001775_060 |
0.1249102523 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000453_290 |
0.1265960455 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 11 |
Hb_142056_010 |
0.1271712013 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002007_330 |
0.1278644848 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 13 |
Hb_002627_010 |
0.1313974017 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 14 |
Hb_000638_230 |
0.1359932558 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000152_520 |
0.1377301017 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 16 |
Hb_010217_020 |
0.1377348135 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 17 |
Hb_002592_070 |
0.1393574335 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 18 |
Hb_004785_170 |
0.1400768339 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 19 |
Hb_009225_070 |
0.1447934971 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 20 |
Hb_002384_030 |
0.1450261618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |