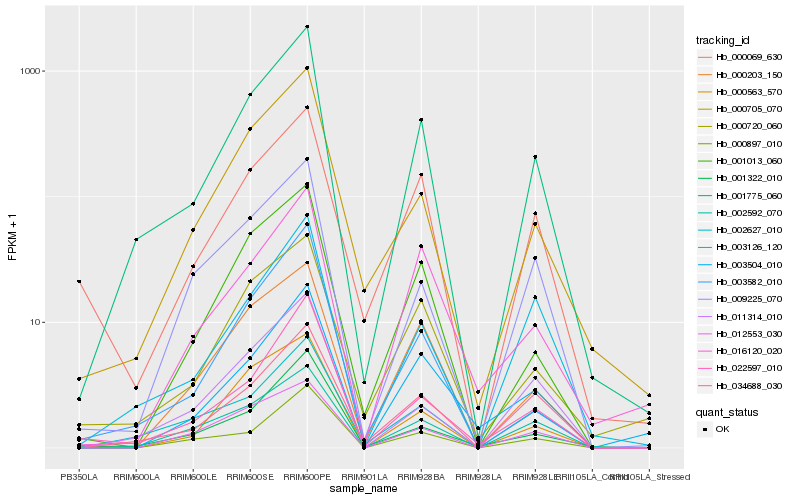
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001775_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_034688_030 |
0.1092817579 |
- |
- |
PREDICTED: uncharacterized protein LOC105642049 [Jatropha curcas] |
| 3 |
Hb_002627_010 |
0.1199981131 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 4 |
Hb_000705_070 |
0.1243724154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003126_120 |
0.1249102523 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 6 |
Hb_002592_070 |
0.1281444641 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 7 |
Hb_003582_010 |
0.1293465984 |
- |
- |
hypothetical protein EUGRSUZ_B026451, partial [Eucalyptus grandis] |
| 8 |
Hb_001013_060 |
0.1303355164 |
- |
- |
- |
| 9 |
Hb_001322_010 |
0.1303878939 |
- |
- |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 10 |
Hb_000720_060 |
0.133764168 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 11 |
Hb_011314_010 |
0.1340693517 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000563_570 |
0.1349365048 |
- |
- |
hypothetical protein JCGZ_18895 [Jatropha curcas] |
| 13 |
Hb_022597_010 |
0.1369201408 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 14 |
Hb_000203_150 |
0.1377789965 |
- |
- |
PREDICTED: uncharacterized protein LOC105116038 [Populus euphratica] |
| 15 |
Hb_000897_010 |
0.1410891961 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 16 |
Hb_000069_630 |
0.1469437179 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 17 |
Hb_003504_010 |
0.1487720604 |
- |
- |
hypothetical protein POPTR_0013s04290g [Populus trichocarpa] |
| 18 |
Hb_012553_030 |
0.1496426892 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 19 |
Hb_009225_070 |
0.1497257508 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 20 |
Hb_016120_020 |
0.1520590372 |
- |
- |
PREDICTED: tetrahydrocannabinolic acid synthase-like [Jatropha curcas] |