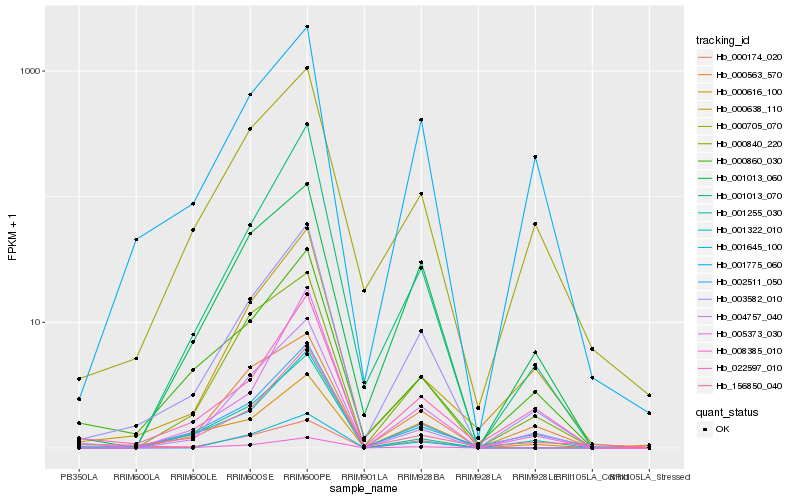
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003582_010 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_B026451, partial [Eucalyptus grandis] |
| 2 |
Hb_008385_010 |
0.109707915 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 3 |
Hb_001013_070 |
0.1117906534 |
- |
- |
- |
| 4 |
Hb_002511_050 |
0.1170002232 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000705_070 |
0.1231965941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001322_010 |
0.1262662376 |
- |
- |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 7 |
Hb_001013_060 |
0.1262811338 |
- |
- |
- |
| 8 |
Hb_001775_060 |
0.1293465984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000840_220 |
0.1304135061 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 10 |
Hb_022597_010 |
0.1334640479 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 11 |
Hb_000860_030 |
0.1367702541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004757_040 |
0.1388651001 |
- |
- |
- |
| 13 |
Hb_000638_110 |
0.1411077585 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0007s04250g [Populus trichocarpa] |
| 14 |
Hb_156850_040 |
0.141673116 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 15 |
Hb_000616_100 |
0.1472085414 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 16 |
Hb_001255_030 |
0.1479212377 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840, partial [Sesamum indicum] |
| 17 |
Hb_000174_020 |
0.1484555365 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 18 |
Hb_001645_100 |
0.1507261273 |
- |
- |
- |
| 19 |
Hb_005373_030 |
0.154168378 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000563_570 |
0.1543970343 |
- |
- |
hypothetical protein JCGZ_18895 [Jatropha curcas] |