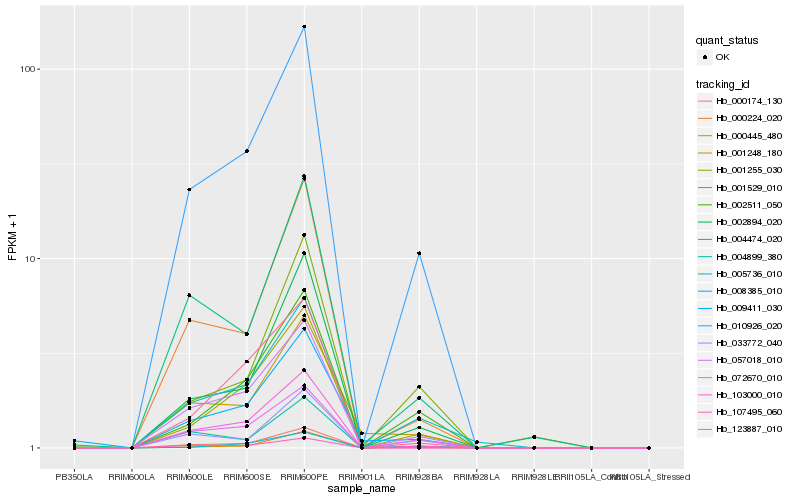
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010926_020 |
0.0 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Camelina sativa] |
| 2 |
Hb_057018_010 |
0.0533178456 |
- |
- |
hypothetical protein JCGZ_04004 [Jatropha curcas] |
| 3 |
Hb_072670_010 |
0.0653281559 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 4 |
Hb_005736_010 |
0.0774829012 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002511_050 |
0.1055872073 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_002894_020 |
0.1076994605 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 7 |
Hb_000224_020 |
0.1142458371 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 8 |
Hb_033772_040 |
0.1223244957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000445_480 |
0.1294597814 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_004474_020 |
0.1307597973 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 11 |
Hb_107495_060 |
0.1310536189 |
- |
- |
Cell division protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001529_010 |
0.132630905 |
- |
- |
- |
| 13 |
Hb_008385_010 |
0.138794485 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 14 |
Hb_001255_030 |
0.140298291 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840, partial [Sesamum indicum] |
| 15 |
Hb_123887_010 |
0.1416726508 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 16 |
Hb_004899_380 |
0.1432683765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001248_180 |
0.1449427595 |
- |
- |
- |
| 18 |
Hb_000174_130 |
0.1459862659 |
- |
- |
flowering locus T-like protein [Fagus crenata] |
| 19 |
Hb_103000_010 |
0.1461891222 |
- |
- |
hypothetical protein POPTR_0007s02210g [Populus trichocarpa] |
| 20 |
Hb_009411_030 |
0.1462713496 |
- |
- |
PREDICTED: bifunctional monodehydroascorbate reductase and carbonic anhydrase nectarin-3-like [Malus domestica] |