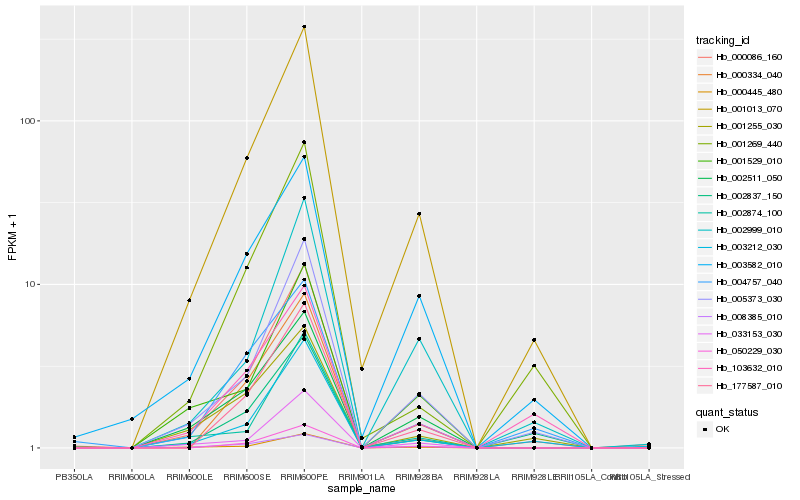
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001013_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_005373_030 |
0.0780930797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003582_010 |
0.1117906534 |
- |
- |
hypothetical protein EUGRSUZ_B026451, partial [Eucalyptus grandis] |
| 4 |
Hb_003212_030 |
0.1142178828 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_033153_030 |
0.1165376633 |
- |
- |
PREDICTED: uncharacterized protein LOC105650145 [Jatropha curcas] |
| 6 |
Hb_050229_030 |
0.118576519 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000445_480 |
0.11928104 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_002999_010 |
0.1211723729 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 9 |
Hb_002511_050 |
0.1227357829 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_008385_010 |
0.1232357792 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 11 |
Hb_177587_010 |
0.1233393744 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 12 |
Hb_000334_040 |
0.1261748396 |
- |
- |
hypothetical protein POPTR_0001s41860g [Populus trichocarpa] |
| 13 |
Hb_001529_010 |
0.1284030048 |
- |
- |
- |
| 14 |
Hb_002837_150 |
0.1334353814 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 15 |
Hb_001269_440 |
0.1340729378 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 16 |
Hb_000086_160 |
0.1357861877 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 17 |
Hb_001255_030 |
0.1408913927 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840, partial [Sesamum indicum] |
| 18 |
Hb_103632_010 |
0.1423194674 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 19 |
Hb_004757_040 |
0.143352579 |
- |
- |
- |
| 20 |
Hb_002874_100 |
0.1434018674 |
- |
- |
cytochrome P450, putative [Ricinus communis] |