| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
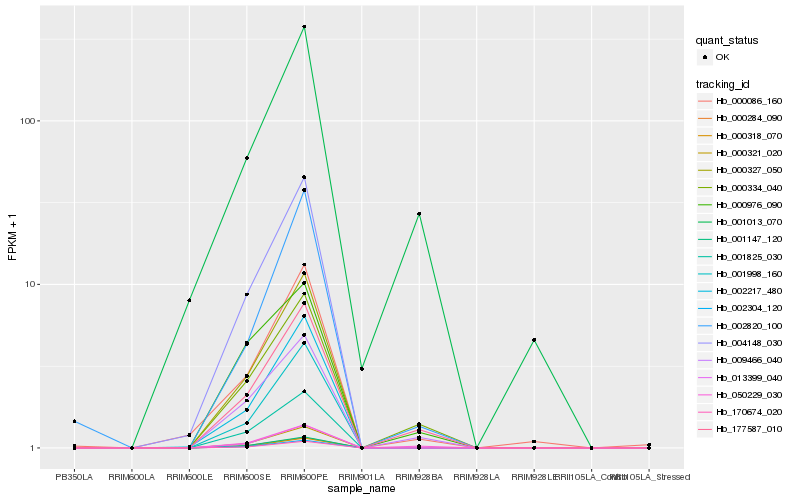
Hb_177587_010 |
0.0 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 2 |
Hb_000334_040 |
0.0306373477 |
- |
- |
hypothetical protein POPTR_0001s41860g [Populus trichocarpa] |
| 3 |
Hb_050229_030 |
0.036362894 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_009466_040 |
0.0596203894 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 5 |
Hb_001013_070 |
0.1233393744 |
- |
- |
- |
| 6 |
Hb_000327_050 |
0.1357542745 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.7-like isoform X1 [Populus euphratica] |
| 7 |
Hb_002304_120 |
0.1357729989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_013399_040 |
0.1360660286 |
- |
- |
hypothetical protein JCGZ_24713 [Jatropha curcas] |
| 9 |
Hb_002820_100 |
0.1360694678 |
- |
- |
hypothetical protein OsI_33031 [Oryza sativa Indica Group] |
| 10 |
Hb_000321_020 |
0.1360866131 |
- |
- |
PREDICTED: uncharacterized protein LOC104891638 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_170674_020 |
0.136963916 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 12 |
Hb_000318_070 |
0.13785221 |
- |
- |
PREDICTED: peroxidase 10-like [Jatropha curcas] |
| 13 |
Hb_001147_120 |
0.1390237434 |
- |
- |
hypothetical protein JCGZ_14802 [Jatropha curcas] |
| 14 |
Hb_000086_160 |
0.1390244788 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 15 |
Hb_000976_090 |
0.1399051206 |
- |
- |
- |
| 16 |
Hb_001825_030 |
0.1399286423 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 17 |
Hb_000284_090 |
0.1400845914 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 18 |
Hb_001998_160 |
0.1416050211 |
- |
- |
PREDICTED: origin recognition complex subunit 1-like [Populus euphratica] |
| 19 |
Hb_004148_030 |
0.1424409514 |
- |
- |
PREDICTED: protein RAFTIN 1B-like [Gossypium raimondii] |
| 20 |
Hb_002217_480 |
0.143586822 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |