| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
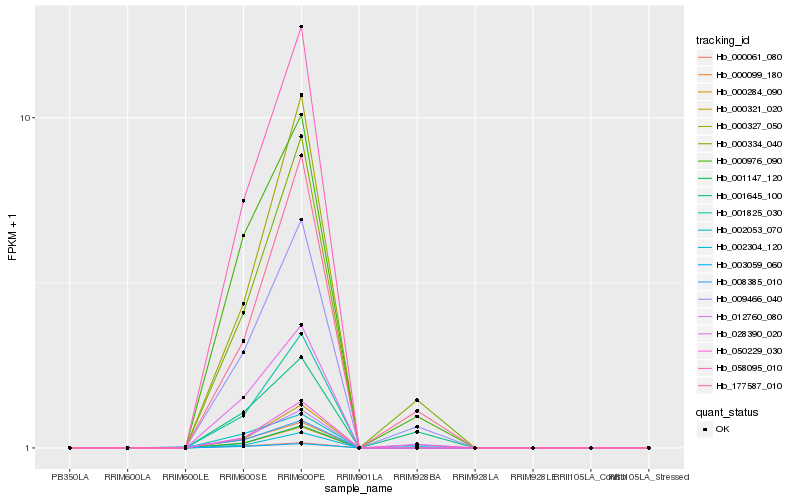
Hb_009466_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 2 |
Hb_000334_040 |
0.0355438341 |
- |
- |
hypothetical protein POPTR_0001s41860g [Populus trichocarpa] |
| 3 |
Hb_177587_010 |
0.0596203894 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 4 |
Hb_050229_030 |
0.0624802698 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_000976_090 |
0.081834878 |
- |
- |
- |
| 6 |
Hb_002053_070 |
0.0857050375 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 14 [Populus euphratica] |
| 7 |
Hb_001645_100 |
0.1189912747 |
- |
- |
- |
| 8 |
Hb_012760_080 |
0.1270358223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_058095_010 |
0.1274832851 |
- |
- |
PREDICTED: uncharacterized protein LOC105109431 [Populus euphratica] |
| 10 |
Hb_000284_090 |
0.1293708599 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 11 |
Hb_001825_030 |
0.1294930121 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 12 |
Hb_001147_120 |
0.1303157108 |
- |
- |
hypothetical protein JCGZ_14802 [Jatropha curcas] |
| 13 |
Hb_000099_180 |
0.1328329113 |
- |
- |
PREDICTED: uncharacterized protein LOC104744055 [Camelina sativa] |
| 14 |
Hb_028390_020 |
0.1340930994 |
- |
- |
- |
| 15 |
Hb_000321_020 |
0.1362258458 |
- |
- |
PREDICTED: uncharacterized protein LOC104891638 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_002304_120 |
0.1385326861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003059_060 |
0.1402870711 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 18 |
Hb_000327_050 |
0.1415910085 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.7-like isoform X1 [Populus euphratica] |
| 19 |
Hb_000061_080 |
0.1428058376 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB28-like [Jatropha curcas] |
| 20 |
Hb_008385_010 |
0.1428462724 |
- |
- |
sulfate transporter, putative [Ricinus communis] |