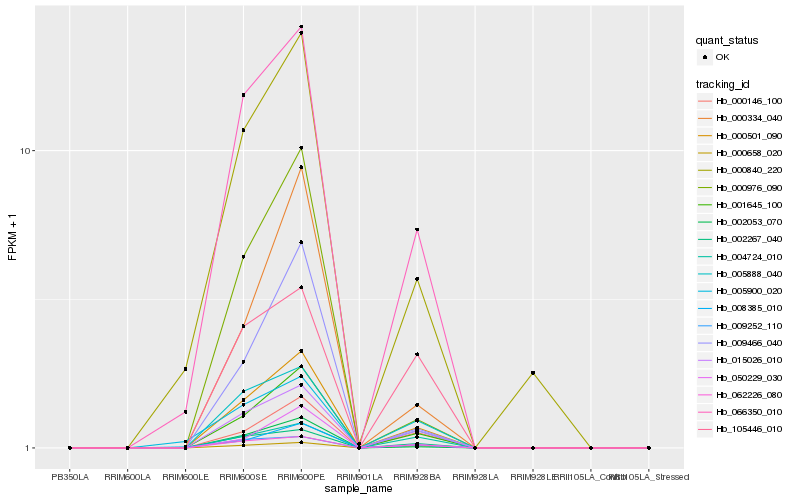
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000501_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_001645_100 |
0.0405168494 |
- |
- |
- |
| 3 |
Hb_015026_010 |
0.0611498732 |
- |
- |
- |
| 4 |
Hb_002267_040 |
0.0853005005 |
- |
- |
- |
| 5 |
Hb_066350_010 |
0.090487071 |
- |
- |
somatic embryogenesis receptor-like kinase 4, partial [Rosa hybrid cultivar] |
| 6 |
Hb_062226_080 |
0.0938700508 |
- |
- |
- |
| 7 |
Hb_005888_040 |
0.1010133354 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Jatropha curcas] |
| 8 |
Hb_000146_100 |
0.108831078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009252_110 |
0.1262133624 |
- |
- |
hypothetical protein JCGZ_11492 [Jatropha curcas] |
| 10 |
Hb_002053_070 |
0.1370289733 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 14 [Populus euphratica] |
| 11 |
Hb_008385_010 |
0.1391776212 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 12 |
Hb_000658_020 |
0.1445308116 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Musa acuminata subsp. malaccensis] |
| 13 |
Hb_009466_040 |
0.1457241041 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 14 |
Hb_004724_010 |
0.1471851621 |
- |
- |
- |
| 15 |
Hb_000976_090 |
0.1494289614 |
- |
- |
- |
| 16 |
Hb_050229_030 |
0.149956037 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_005900_020 |
0.1508426006 |
- |
- |
- |
| 18 |
Hb_000334_040 |
0.1516547438 |
- |
- |
hypothetical protein POPTR_0001s41860g [Populus trichocarpa] |
| 19 |
Hb_000840_220 |
0.1536015772 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 20 |
Hb_105446_010 |
0.1564750749 |
- |
- |
- |