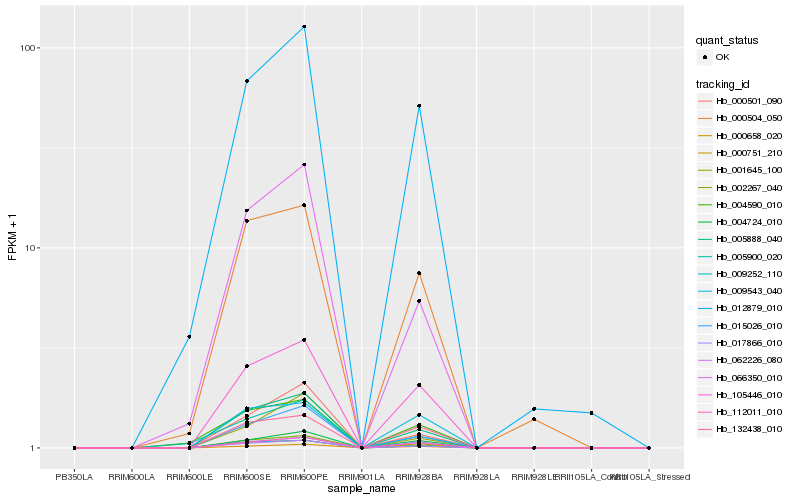
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005888_040 |
0.0 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Jatropha curcas] |
| 2 |
Hb_062226_080 |
0.0082875985 |
- |
- |
- |
| 3 |
Hb_002267_040 |
0.0251660878 |
- |
- |
- |
| 4 |
Hb_009252_110 |
0.0341856135 |
- |
- |
hypothetical protein JCGZ_11492 [Jatropha curcas] |
| 5 |
Hb_015026_010 |
0.0445268259 |
- |
- |
- |
| 6 |
Hb_105446_010 |
0.0754867136 |
- |
- |
- |
| 7 |
Hb_066350_010 |
0.0820436289 |
- |
- |
somatic embryogenesis receptor-like kinase 4, partial [Rosa hybrid cultivar] |
| 8 |
Hb_000658_020 |
0.0995601766 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Musa acuminata subsp. malaccensis] |
| 9 |
Hb_000501_090 |
0.1010133354 |
- |
- |
- |
| 10 |
Hb_112011_010 |
0.1079917705 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_004724_010 |
0.1081398007 |
- |
- |
- |
| 12 |
Hb_000751_210 |
0.1091926131 |
- |
- |
PREDICTED: petal death protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_012879_010 |
0.1166284205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000504_050 |
0.1173999416 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 15 |
Hb_017866_010 |
0.1269978474 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 16 |
Hb_132438_010 |
0.1274277899 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 17 |
Hb_001645_100 |
0.1385090882 |
- |
- |
- |
| 18 |
Hb_009543_040 |
0.1456868335 |
- |
- |
- |
| 19 |
Hb_005900_020 |
0.1474995244 |
- |
- |
- |
| 20 |
Hb_004590_010 |
0.1479238204 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |