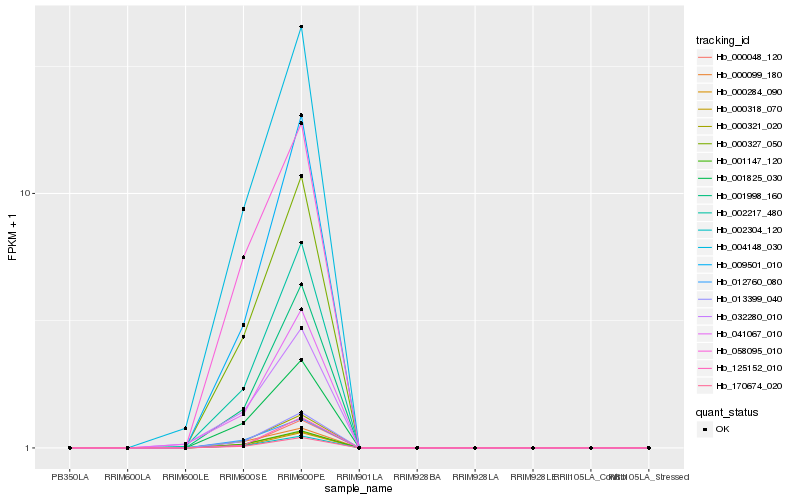
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_050 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.7-like isoform X1 [Populus euphratica] |
| 2 |
Hb_013399_040 |
0.0065453149 |
- |
- |
hypothetical protein JCGZ_24713 [Jatropha curcas] |
| 3 |
Hb_002304_120 |
0.0073492103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000321_020 |
0.0134754483 |
- |
- |
PREDICTED: uncharacterized protein LOC104891638 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_170674_020 |
0.0153082078 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 6 |
Hb_000318_070 |
0.0210856144 |
- |
- |
PREDICTED: peroxidase 10-like [Jatropha curcas] |
| 7 |
Hb_001147_120 |
0.0337612455 |
- |
- |
hypothetical protein JCGZ_14802 [Jatropha curcas] |
| 8 |
Hb_001998_160 |
0.0374673325 |
- |
- |
PREDICTED: origin recognition complex subunit 1-like [Populus euphratica] |
| 9 |
Hb_001825_030 |
0.0377285955 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 10 |
Hb_000284_090 |
0.0383693998 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 11 |
Hb_004148_030 |
0.0444763394 |
- |
- |
PREDICTED: protein RAFTIN 1B-like [Gossypium raimondii] |
| 12 |
Hb_002217_480 |
0.045119904 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 13 |
Hb_009501_010 |
0.057808036 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 14 |
Hb_012760_080 |
0.0613097436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_058095_010 |
0.0739009436 |
- |
- |
PREDICTED: uncharacterized protein LOC105109431 [Populus euphratica] |
| 16 |
Hb_041067_010 |
0.0796967625 |
- |
- |
hypothetical protein JCGZ_00780 [Jatropha curcas] |
| 17 |
Hb_032280_010 |
0.0917971268 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 18 |
Hb_125152_010 |
0.0956602671 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 19 |
Hb_000048_120 |
0.096973417 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 20 |
Hb_000099_180 |
0.1020216428 |
- |
- |
PREDICTED: uncharacterized protein LOC104744055 [Camelina sativa] |