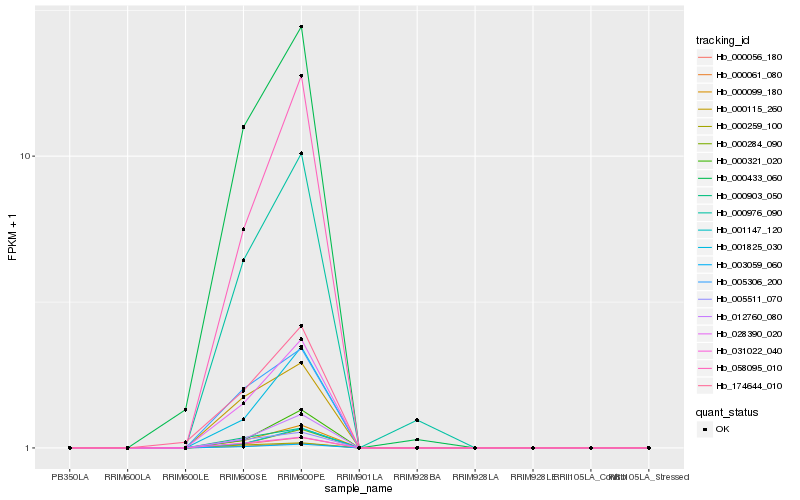
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003059_060 |
0.0 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 2 |
Hb_000061_080 |
0.005777267 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB28-like [Jatropha curcas] |
| 3 |
Hb_000056_180 |
0.0098241352 |
- |
- |
- |
| 4 |
Hb_028390_020 |
0.0167503099 |
- |
- |
- |
| 5 |
Hb_000099_180 |
0.0209006831 |
- |
- |
PREDICTED: uncharacterized protein LOC104744055 [Camelina sativa] |
| 6 |
Hb_031022_040 |
0.0460495682 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_058095_010 |
0.0491683709 |
- |
- |
PREDICTED: uncharacterized protein LOC105109431 [Populus euphratica] |
| 8 |
Hb_012760_080 |
0.0617815557 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000903_050 |
0.0715199478 |
- |
- |
hypothetical protein A259_26892 [Pseudomonas syringae pv. actinidiae ICMP 19070] |
| 10 |
Hb_005306_200 |
0.0742658528 |
- |
- |
- |
| 11 |
Hb_005511_070 |
0.0787356458 |
- |
- |
- |
| 12 |
Hb_000115_260 |
0.0803207809 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH75 [Vitis vinifera] |
| 13 |
Hb_000284_090 |
0.084684259 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 14 |
Hb_001825_030 |
0.0853224804 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 15 |
Hb_000433_060 |
0.0867120039 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 16 |
Hb_001147_120 |
0.0892718988 |
- |
- |
hypothetical protein JCGZ_14802 [Jatropha curcas] |
| 17 |
Hb_000976_090 |
0.0992512866 |
- |
- |
- |
| 18 |
Hb_174644_010 |
0.1022600121 |
- |
- |
- |
| 19 |
Hb_000259_100 |
0.1022897732 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 20 |
Hb_000321_020 |
0.1094115225 |
- |
- |
PREDICTED: uncharacterized protein LOC104891638 [Beta vulgaris subsp. vulgaris] |