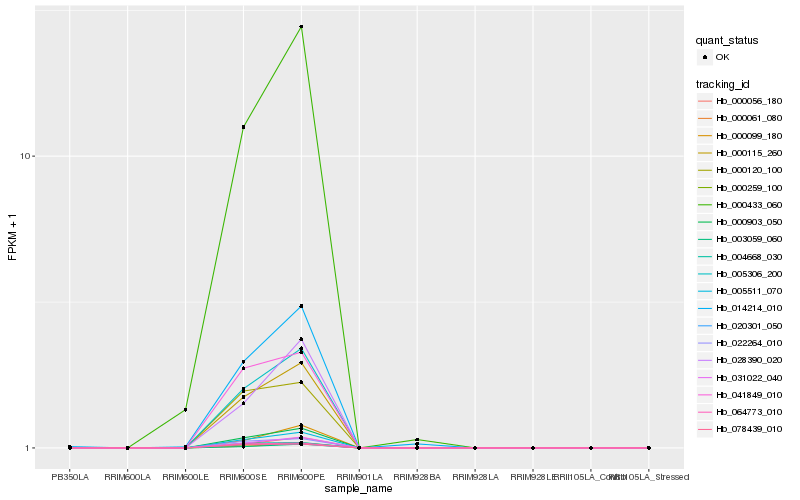
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_260 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH75 [Vitis vinifera] |
| 2 |
Hb_005511_070 |
0.0015897984 |
- |
- |
- |
| 3 |
Hb_005306_200 |
0.006071813 |
- |
- |
- |
| 4 |
Hb_000903_050 |
0.0088245401 |
- |
- |
hypothetical protein A259_26892 [Pseudomonas syringae pv. actinidiae ICMP 19070] |
| 5 |
Hb_000259_100 |
0.0220508534 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 6 |
Hb_031022_040 |
0.0343323976 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_022264_010 |
0.065512734 |
- |
- |
PREDICTED: uncharacterized protein LOC105795741 [Gossypium raimondii] |
| 8 |
Hb_000056_180 |
0.0705246787 |
- |
- |
- |
| 9 |
Hb_000061_080 |
0.0745610321 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB28-like [Jatropha curcas] |
| 10 |
Hb_003059_060 |
0.0803207809 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 11 |
Hb_000433_060 |
0.0815399888 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 12 |
Hb_041849_010 |
0.0844426631 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 13 |
Hb_014214_010 |
0.0930186767 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_064773_010 |
0.0967280856 |
- |
- |
PREDICTED: uncharacterized protein LOC103699620, partial [Phoenix dactylifera] |
| 15 |
Hb_028390_020 |
0.0970030685 |
- |
- |
- |
| 16 |
Hb_000120_100 |
0.0997443059 |
- |
- |
- |
| 17 |
Hb_000099_180 |
0.1011324917 |
- |
- |
PREDICTED: uncharacterized protein LOC104744055 [Camelina sativa] |
| 18 |
Hb_004668_030 |
0.1063989103 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 19 |
Hb_020301_050 |
0.1069265757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_078439_010 |
0.1101044141 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |