| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
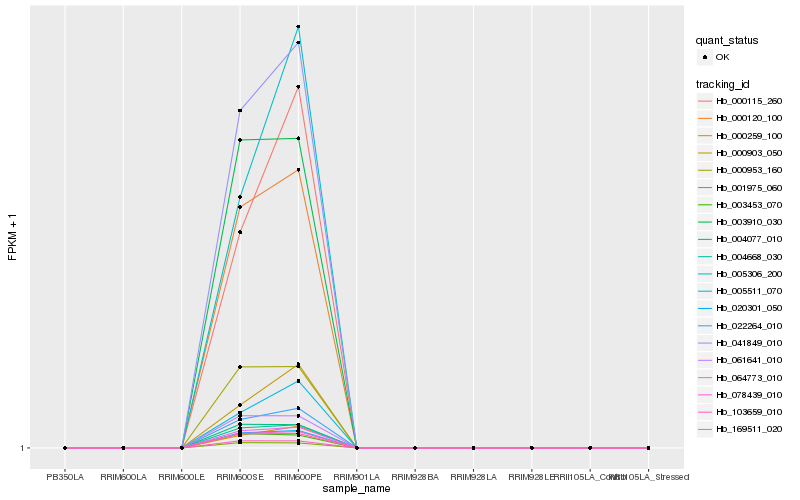
Hb_022264_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105795741 [Gossypium raimondii] |
| 2 |
Hb_041849_010 |
0.0189695463 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 3 |
Hb_064773_010 |
0.031289636 |
- |
- |
PREDICTED: uncharacterized protein LOC103699620, partial [Phoenix dactylifera] |
| 4 |
Hb_000120_100 |
0.0343154271 |
- |
- |
- |
| 5 |
Hb_004668_030 |
0.0409926104 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 6 |
Hb_020301_050 |
0.0415221519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000259_100 |
0.0434866665 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 8 |
Hb_078439_010 |
0.0447115547 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 9 |
Hb_000115_260 |
0.065512734 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH75 [Vitis vinifera] |
| 10 |
Hb_169511_020 |
0.067078145 |
- |
- |
PREDICTED: uncharacterized protein LOC105037523, partial [Elaeis guineensis] |
| 11 |
Hb_005511_070 |
0.0670997223 |
- |
- |
- |
| 12 |
Hb_003910_030 |
0.0713991445 |
- |
- |
PREDICTED: probable disease resistance protein At5g66910 [Populus euphratica] |
| 13 |
Hb_005306_200 |
0.0715730564 |
- |
- |
- |
| 14 |
Hb_000953_160 |
0.0718020598 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 15 |
Hb_061641_010 |
0.0733153124 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 16 |
Hb_000903_050 |
0.0743198931 |
- |
- |
hypothetical protein A259_26892 [Pseudomonas syringae pv. actinidiae ICMP 19070] |
| 17 |
Hb_004077_010 |
0.0759648033 |
- |
- |
retinoblastoma-binding protein, putative [Ricinus communis] |
| 18 |
Hb_103659_010 |
0.0768286047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001975_060 |
0.0841895614 |
- |
- |
hypothetical protein JCGZ_11079 [Jatropha curcas] |
| 20 |
Hb_003453_070 |
0.0956527927 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor LEP [Jatropha curcas] |