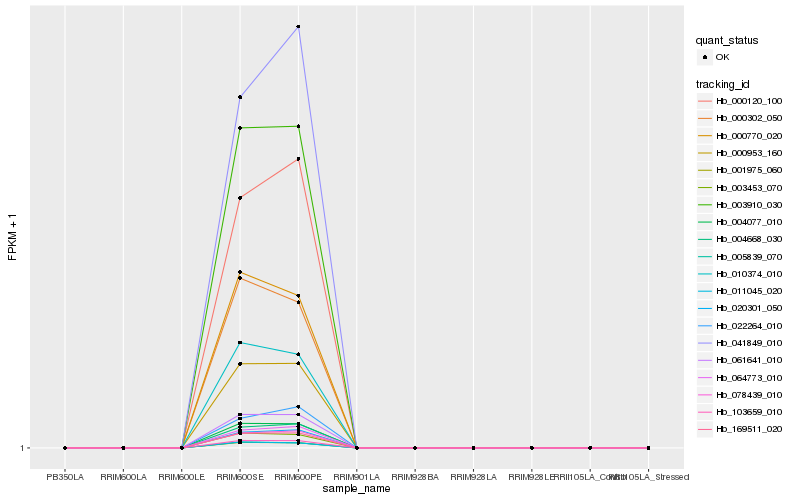
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_078439_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 2 |
Hb_020301_050 |
0.003191501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004668_030 |
0.0037213601 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 4 |
Hb_000120_100 |
0.0104017928 |
- |
- |
- |
| 5 |
Hb_064773_010 |
0.0134285944 |
- |
- |
PREDICTED: uncharacterized protein LOC103699620, partial [Phoenix dactylifera] |
| 6 |
Hb_169511_020 |
0.0223901937 |
- |
- |
PREDICTED: uncharacterized protein LOC105037523, partial [Elaeis guineensis] |
| 7 |
Hb_041849_010 |
0.0257498039 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 8 |
Hb_003910_030 |
0.0267175362 |
- |
- |
PREDICTED: probable disease resistance protein At5g66910 [Populus euphratica] |
| 9 |
Hb_000953_160 |
0.0271210723 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 10 |
Hb_061641_010 |
0.0286367011 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 11 |
Hb_004077_010 |
0.0312905217 |
- |
- |
retinoblastoma-binding protein, putative [Ricinus communis] |
| 12 |
Hb_103659_010 |
0.0321557814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001975_060 |
0.0395300951 |
- |
- |
hypothetical protein JCGZ_11079 [Jatropha curcas] |
| 14 |
Hb_022264_010 |
0.0447115547 |
- |
- |
PREDICTED: uncharacterized protein LOC105795741 [Gossypium raimondii] |
| 15 |
Hb_003453_070 |
0.051017438 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor LEP [Jatropha curcas] |
| 16 |
Hb_005839_070 |
0.0552271403 |
- |
- |
hypothetical protein POPTR_0013s08160g [Populus trichocarpa] |
| 17 |
Hb_010374_010 |
0.0557160004 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 18 |
Hb_000770_020 |
0.0632022313 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 19 |
Hb_011045_020 |
0.0643236087 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 20 |
Hb_000302_050 |
0.0654604727 |
- |
- |
- |