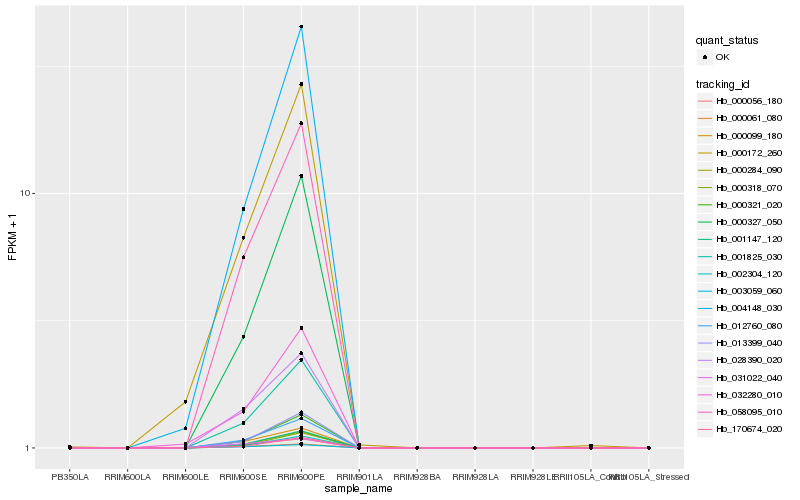
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_058095_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105109431 [Populus euphratica] |
| 2 |
Hb_012760_080 |
0.0126390726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000099_180 |
0.02828605 |
- |
- |
PREDICTED: uncharacterized protein LOC104744055 [Camelina sativa] |
| 4 |
Hb_028390_020 |
0.0324348265 |
- |
- |
- |
| 5 |
Hb_000284_090 |
0.03561974 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 6 |
Hb_001825_030 |
0.0362607405 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 7 |
Hb_001147_120 |
0.0402281215 |
- |
- |
hypothetical protein JCGZ_14802 [Jatropha curcas] |
| 8 |
Hb_003059_060 |
0.0491683709 |
- |
- |
hypothetical protein CICLE_v100140161mg, partial [Citrus clementina] |
| 9 |
Hb_000061_080 |
0.0549361767 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB28-like [Jatropha curcas] |
| 10 |
Hb_000056_180 |
0.0589753399 |
- |
- |
- |
| 11 |
Hb_000321_020 |
0.0604806869 |
- |
- |
PREDICTED: uncharacterized protein LOC104891638 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_002304_120 |
0.066585272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000327_050 |
0.0739009436 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.7-like isoform X1 [Populus euphratica] |
| 14 |
Hb_004148_030 |
0.0770011865 |
- |
- |
PREDICTED: protein RAFTIN 1B-like [Gossypium raimondii] |
| 15 |
Hb_013399_040 |
0.0804091836 |
- |
- |
hypothetical protein JCGZ_24713 [Jatropha curcas] |
| 16 |
Hb_170674_020 |
0.0891113654 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 17 |
Hb_000318_070 |
0.0948415389 |
- |
- |
PREDICTED: peroxidase 10-like [Jatropha curcas] |
| 18 |
Hb_031022_040 |
0.0950949482 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_032280_010 |
0.0966102937 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000172_260 |
0.0985935751 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |