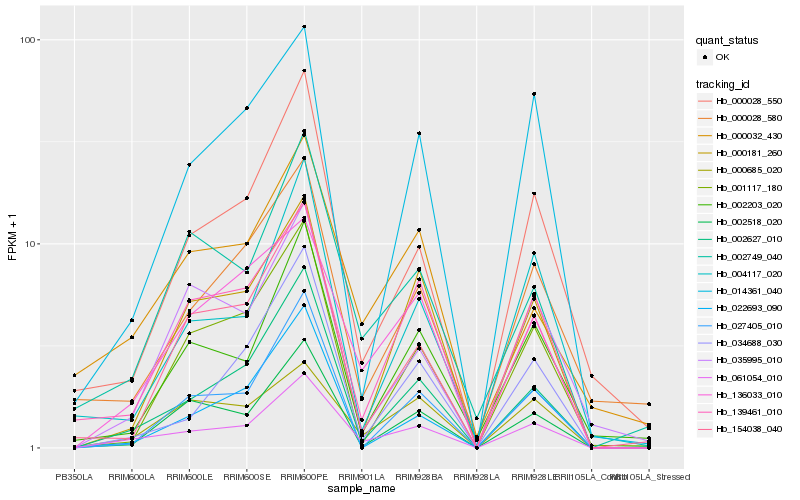
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061054_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_01675 [Jatropha curcas] |
| 2 |
Hb_002627_010 |
0.119112617 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 3 |
Hb_136033_010 |
0.1534177991 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Gossypium raimondii] |
| 4 |
Hb_000028_550 |
0.1535642996 |
- |
- |
unknown [Medicago truncatula] |
| 5 |
Hb_139461_010 |
0.1615895991 |
desease resistance |
Gene Name: NB-ARC |
Cc-nbs-lrr resistance protein, putative [Theobroma cacao] |
| 6 |
Hb_002749_040 |
0.1626394123 |
- |
- |
Ribosomal protein L6 family protein [Theobroma cacao] |
| 7 |
Hb_000181_260 |
0.1644566448 |
- |
- |
PREDICTED: uncharacterized protein LOC105139156 [Populus euphratica] |
| 8 |
Hb_027405_010 |
0.1648457791 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 9 |
Hb_002203_020 |
0.1649961657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000685_020 |
0.1657026646 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 11 |
Hb_034688_030 |
0.1657170616 |
- |
- |
PREDICTED: uncharacterized protein LOC105642049 [Jatropha curcas] |
| 12 |
Hb_154038_040 |
0.167477176 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 13 |
Hb_002518_020 |
0.1695449383 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 14 |
Hb_014361_040 |
0.1700344595 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 15 |
Hb_004117_020 |
0.1721382418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001117_180 |
0.1741571324 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 17 |
Hb_022693_090 |
0.1769134606 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000028_580 |
0.1799432808 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 19 |
Hb_035995_010 |
0.1812455675 |
- |
- |
- |
| 20 |
Hb_000032_430 |
0.1813645536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |