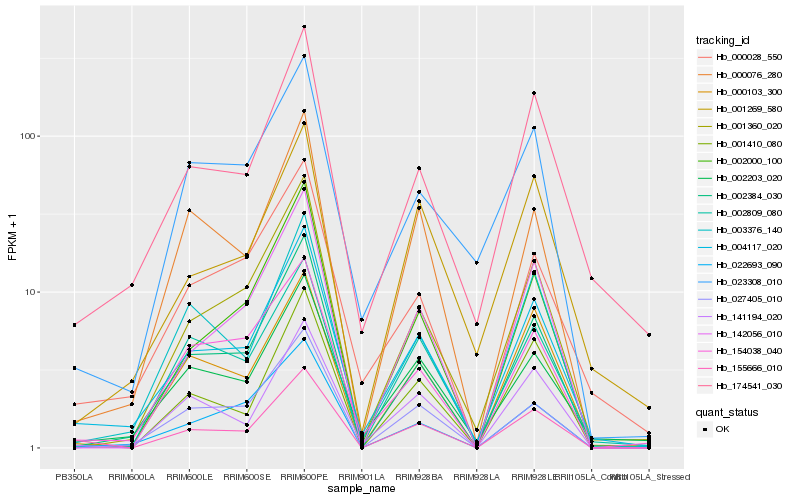
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004117_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_174541_030 |
0.0891243105 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 3 |
Hb_002203_020 |
0.0942876924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002384_030 |
0.1085754412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001360_020 |
0.1089725157 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 6 |
Hb_000028_550 |
0.1128284415 |
- |
- |
unknown [Medicago truncatula] |
| 7 |
Hb_155666_010 |
0.1151875495 |
- |
- |
hypothetical protein JCGZ_01214 [Jatropha curcas] |
| 8 |
Hb_003376_140 |
0.1231780033 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 9 |
Hb_002809_080 |
0.1288133161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_154038_040 |
0.1309271389 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 11 |
Hb_141194_020 |
0.1311875912 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 12 |
Hb_002000_100 |
0.1312008386 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 13 |
Hb_000076_280 |
0.1314683486 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 14 |
Hb_001269_580 |
0.1324544607 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 15 |
Hb_023308_010 |
0.1333273939 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 16 |
Hb_000103_300 |
0.1338006368 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 17 |
Hb_142056_010 |
0.1356958679 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_027405_010 |
0.1367396412 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 19 |
Hb_001410_080 |
0.1367924688 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_022693_090 |
0.1385995634 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |