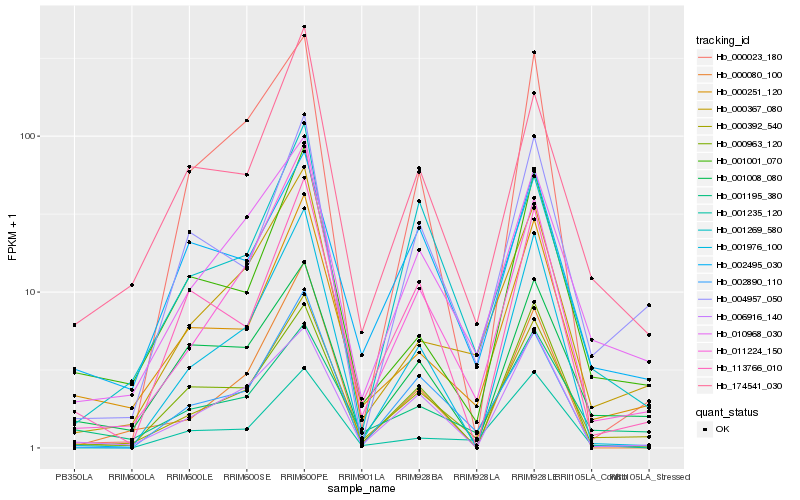
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_540 |
0.0 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Sesamum indicum] |
| 2 |
Hb_010968_030 |
0.0953961277 |
- |
- |
PREDICTED: uncharacterized protein LOC105643149 [Jatropha curcas] |
| 3 |
Hb_011224_150 |
0.0990844509 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000251_120 |
0.1090927293 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000367_080 |
0.1101460466 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 6 |
Hb_004957_050 |
0.1211056721 |
- |
- |
hypothetical protein POPTR_0010s12360g [Populus trichocarpa] |
| 7 |
Hb_002890_110 |
0.121852802 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 8 |
Hb_001976_100 |
0.1226630641 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_174541_030 |
0.132550059 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 10 |
Hb_006916_140 |
0.1334387446 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 11 |
Hb_001001_070 |
0.1360657853 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 12 |
Hb_113766_010 |
0.1416840905 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |
| 13 |
Hb_001195_380 |
0.1417820472 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 14 |
Hb_001269_580 |
0.1438674972 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 15 |
Hb_001008_080 |
0.1471510886 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 16 |
Hb_000080_100 |
0.1481473071 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 17 |
Hb_000963_120 |
0.1494980474 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002495_030 |
0.1518699724 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000023_180 |
0.1529678462 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 20 |
Hb_001235_120 |
0.1533922907 |
- |
- |
hypothetical protein JCGZ_03395 [Jatropha curcas] |