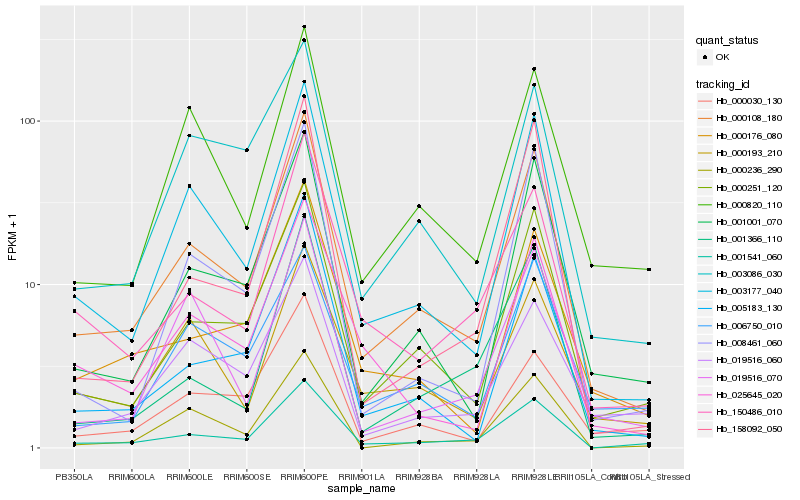
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000108_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 2 |
Hb_003177_040 |
0.0766245568 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 3 |
Hb_000251_120 |
0.0809705598 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 4 |
Hb_001001_070 |
0.0979171061 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 5 |
Hb_001541_060 |
0.1090463477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000176_080 |
0.1214384515 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |
| 7 |
Hb_005183_130 |
0.1265271703 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 8 |
Hb_000236_290 |
0.1272434222 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 9 |
Hb_150486_010 |
0.1273171485 |
- |
- |
hypothetical protein CICLE_v100116733mg, partial [Citrus clementina] |
| 10 |
Hb_003086_030 |
0.1298268667 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 11 |
Hb_000820_110 |
0.1301841589 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 12 |
Hb_025645_020 |
0.1315745517 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 13 |
Hb_008461_060 |
0.1322903147 |
- |
- |
hypothetical protein JCGZ_17379 [Jatropha curcas] |
| 14 |
Hb_019516_060 |
0.1377915399 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 15 |
Hb_001366_110 |
0.1383809308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000193_210 |
0.1395517197 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 17 |
Hb_000030_130 |
0.1410683336 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_006750_010 |
0.1434401039 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 19 |
Hb_158092_050 |
0.1435856441 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 20 |
Hb_019516_070 |
0.1440553033 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |