| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
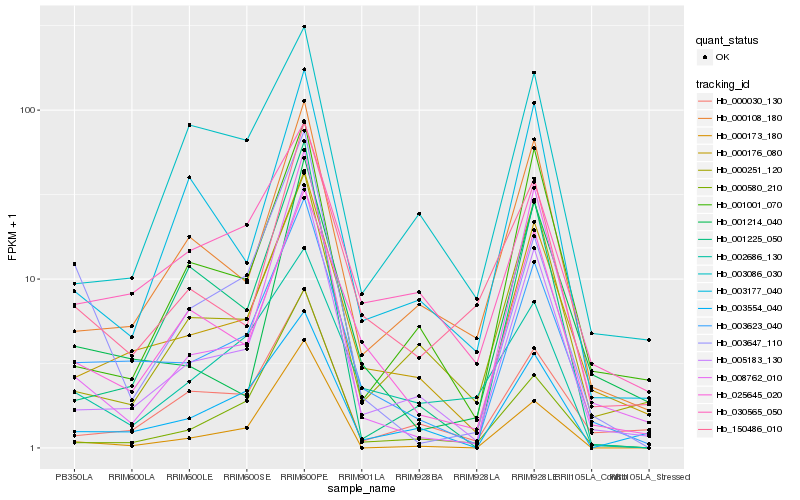
Hb_003623_040 |
0.0 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_000176_080 |
0.0958881093 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |
| 3 |
Hb_005183_130 |
0.1391419335 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 4 |
Hb_025645_020 |
0.1396733299 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 5 |
Hb_030565_050 |
0.1558965331 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 6 |
Hb_003554_040 |
0.1643906545 |
- |
- |
- |
| 7 |
Hb_000108_180 |
0.172482696 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 8 |
Hb_150486_010 |
0.1762546923 |
- |
- |
hypothetical protein CICLE_v100116733mg, partial [Citrus clementina] |
| 9 |
Hb_000030_130 |
0.177965871 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_001214_040 |
0.1792941968 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 11 |
Hb_003647_110 |
0.179377107 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 12 |
Hb_002686_130 |
0.1900421444 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105650401 [Jatropha curcas] |
| 13 |
Hb_001001_070 |
0.1956180115 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 14 |
Hb_000580_210 |
0.1965677783 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 15 [Jatropha curcas] |
| 15 |
Hb_000173_180 |
0.1975305522 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000251_120 |
0.1977572811 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001225_050 |
0.1980628855 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 18 |
Hb_003177_040 |
0.1980877644 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 19 |
Hb_008762_010 |
0.201859263 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 20 |
Hb_003086_030 |
0.2065416599 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |