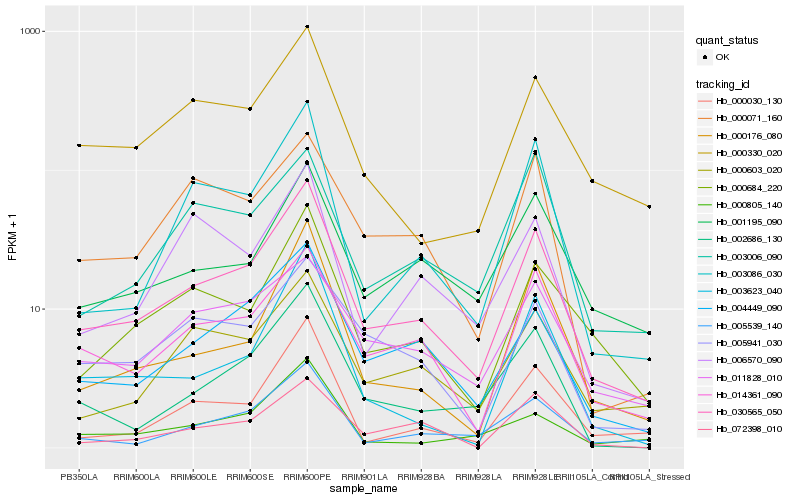
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030565_050 |
0.0 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 2 |
Hb_014361_090 |
0.115317864 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 3 |
Hb_004449_090 |
0.1159595713 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 4 |
Hb_000684_220 |
0.1301496007 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_005539_140 |
0.1357807315 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 6 |
Hb_000176_080 |
0.1367556323 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |
| 7 |
Hb_000603_020 |
0.1387355341 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003086_030 |
0.1390840449 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 9 |
Hb_002686_130 |
0.1404596336 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105650401 [Jatropha curcas] |
| 10 |
Hb_001195_090 |
0.1424253421 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 11 |
Hb_000330_020 |
0.1440697954 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 12 |
Hb_005941_030 |
0.1482330159 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000030_130 |
0.1484061324 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_011828_010 |
0.1507239299 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 15 |
Hb_003623_040 |
0.1558965331 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_000805_140 |
0.1608485892 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 17 |
Hb_006570_090 |
0.1621822951 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 18 |
Hb_000071_160 |
0.1624743142 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 19 |
Hb_072398_010 |
0.1626054527 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 20 |
Hb_003006_090 |
0.1639842405 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |