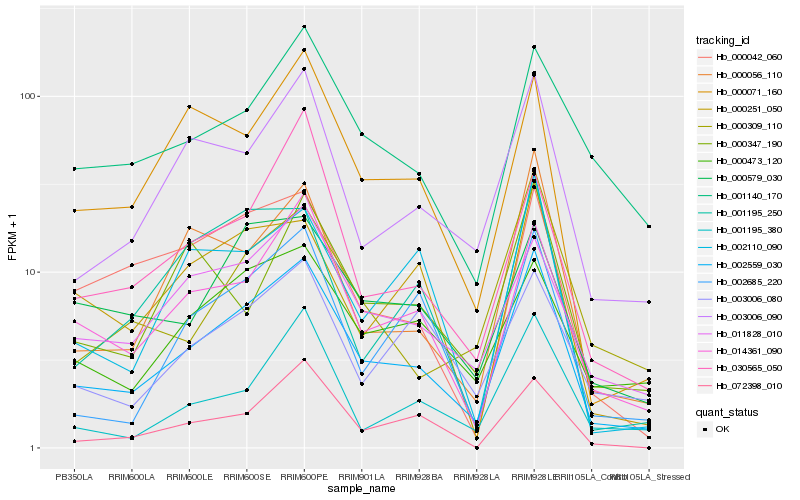
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002559_030 |
0.0 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 2 |
Hb_014361_090 |
0.1240366733 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 3 |
Hb_003006_090 |
0.1313667652 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 4 |
Hb_011828_010 |
0.1437139656 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 5 |
Hb_000056_110 |
0.1448585993 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 6 |
Hb_000071_160 |
0.156261079 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 7 |
Hb_002685_220 |
0.1578258367 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000473_120 |
0.1581067199 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 9 |
Hb_072398_010 |
0.1632646595 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 10 |
Hb_001195_380 |
0.1637461703 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 11 |
Hb_002110_090 |
0.1642848086 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 12 |
Hb_001140_170 |
0.1643012442 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 13 |
Hb_000309_110 |
0.1643398778 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 14 |
Hb_000251_050 |
0.1650623643 |
- |
- |
hypothetical protein JCGZ_21678 [Jatropha curcas] |
| 15 |
Hb_000579_030 |
0.1654027114 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 16 |
Hb_000347_190 |
0.1687847267 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 17 |
Hb_001195_250 |
0.1695181336 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000042_060 |
0.1709820172 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 19 |
Hb_030565_050 |
0.171235268 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 20 |
Hb_003006_080 |
0.1719730815 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |