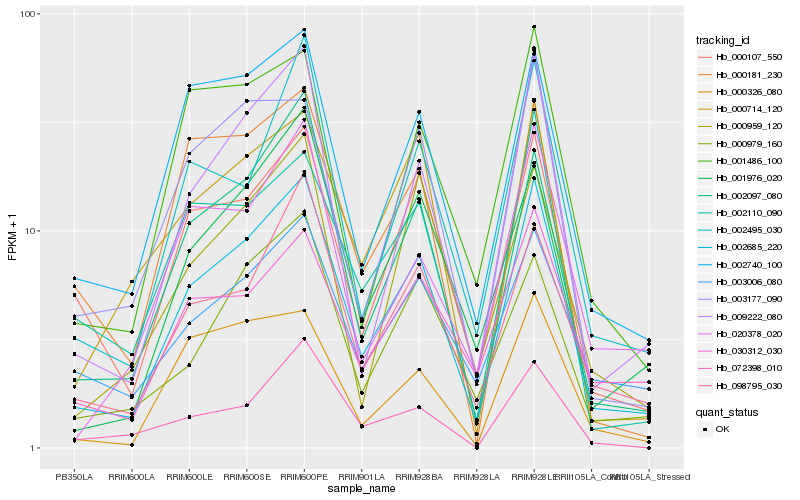
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002685_220 |
0.0 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_009222_080 |
0.0999609458 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 3 |
Hb_002097_080 |
0.1080066131 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 4 |
Hb_000107_550 |
0.1178350243 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 5 |
Hb_002740_100 |
0.1210526866 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 6 |
Hb_002495_030 |
0.123633244 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001976_020 |
0.12531115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_098795_030 |
0.1257856665 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 9 |
Hb_001486_100 |
0.1357227243 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 10 |
Hb_020378_020 |
0.1370164196 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000979_160 |
0.1385437796 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 12 |
Hb_002110_090 |
0.1386648574 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 13 |
Hb_003177_090 |
0.1398836662 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 14 |
Hb_000326_080 |
0.1405568456 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000714_120 |
0.142287928 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 16 |
Hb_030312_030 |
0.1436557341 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003006_080 |
0.143676562 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_072398_010 |
0.144715293 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 19 |
Hb_000181_230 |
0.1467603077 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_000959_120 |
0.147022127 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |