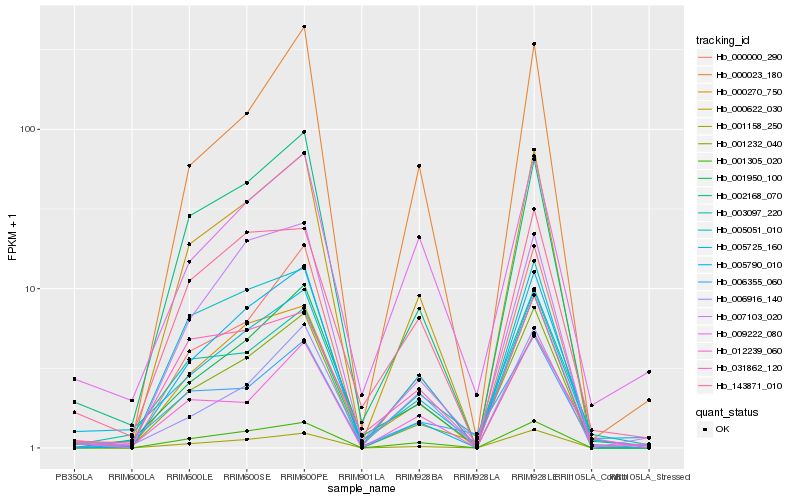
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005725_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 2 |
Hb_001950_100 |
0.1030644857 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 3 |
Hb_001232_040 |
0.113848835 |
- |
- |
PREDICTED: uncharacterized protein LOC105634974 [Jatropha curcas] |
| 4 |
Hb_009222_080 |
0.1162389207 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 5 |
Hb_003097_220 |
0.1185627855 |
- |
- |
hypothetical protein POPTR_0001s06150g [Populus trichocarpa] |
| 6 |
Hb_000622_030 |
0.1270539993 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 7 |
Hb_002168_070 |
0.129436782 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_143871_010 |
0.1344095378 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 9 |
Hb_006355_060 |
0.1378700545 |
- |
- |
PREDICTED: transcription factor PIF1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_005790_010 |
0.1401484561 |
- |
- |
hypothetical protein POPTR_0004s24080g [Populus trichocarpa] |
| 11 |
Hb_006916_140 |
0.1425485135 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 12 |
Hb_001158_250 |
0.1441221233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001305_020 |
0.1444127476 |
- |
- |
PREDICTED: uncharacterized protein LOC102623247 [Citrus sinensis] |
| 14 |
Hb_007103_020 |
0.1468408928 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |
| 15 |
Hb_000000_290 |
0.1470779575 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 16 |
Hb_031862_120 |
0.1490680394 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 17 |
Hb_012239_060 |
0.1503389446 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial [Jatropha curcas] |
| 18 |
Hb_005051_010 |
0.1517434758 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000023_180 |
0.1522477396 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 20 |
Hb_000270_750 |
0.152576045 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |