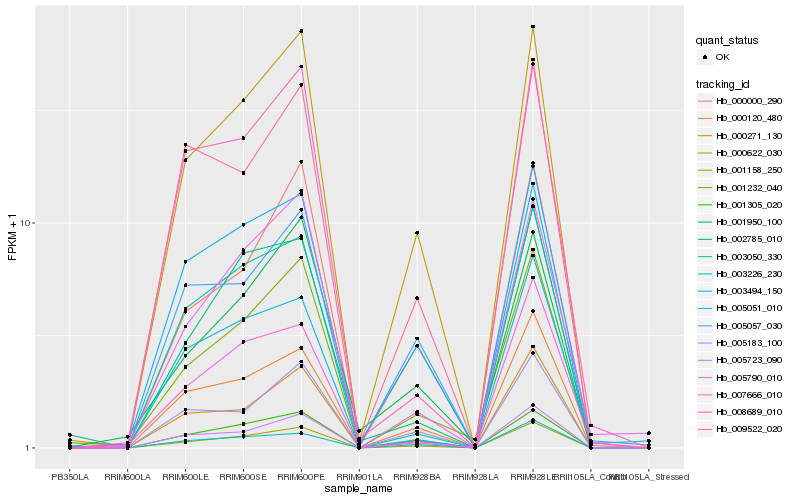
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001158_250 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000622_030 |
0.050063201 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 3 |
Hb_000120_480 |
0.0721901531 |
- |
- |
- |
| 4 |
Hb_005723_090 |
0.0724773653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001232_040 |
0.079508287 |
- |
- |
PREDICTED: uncharacterized protein LOC105634974 [Jatropha curcas] |
| 6 |
Hb_001305_020 |
0.0844110661 |
- |
- |
PREDICTED: uncharacterized protein LOC102623247 [Citrus sinensis] |
| 7 |
Hb_000271_130 |
0.091174482 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 8 |
Hb_003050_330 |
0.0935701023 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 9 |
Hb_005183_100 |
0.0962981433 |
- |
- |
hypothetical protein CICLE_v10002898mg [Citrus clementina] |
| 10 |
Hb_000000_290 |
0.0970610614 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 11 |
Hb_003226_230 |
0.1015889744 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 12 |
Hb_005051_010 |
0.103310802 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_005790_010 |
0.1033690597 |
- |
- |
hypothetical protein POPTR_0004s24080g [Populus trichocarpa] |
| 14 |
Hb_002785_010 |
0.1049881464 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 15 |
Hb_005057_030 |
0.1122130279 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 16 |
Hb_009522_020 |
0.1131435926 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 17 |
Hb_008689_010 |
0.1187573901 |
- |
- |
hypothetical protein POPTR_0006s25330g [Populus trichocarpa] |
| 18 |
Hb_001950_100 |
0.1193144983 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 19 |
Hb_007666_010 |
0.1215983951 |
- |
- |
PREDICTED: uncharacterized protein LOC105649266 [Jatropha curcas] |
| 20 |
Hb_003494_150 |
0.1239037891 |
- |
- |
Silicon transporter, putative [Ricinus communis] |