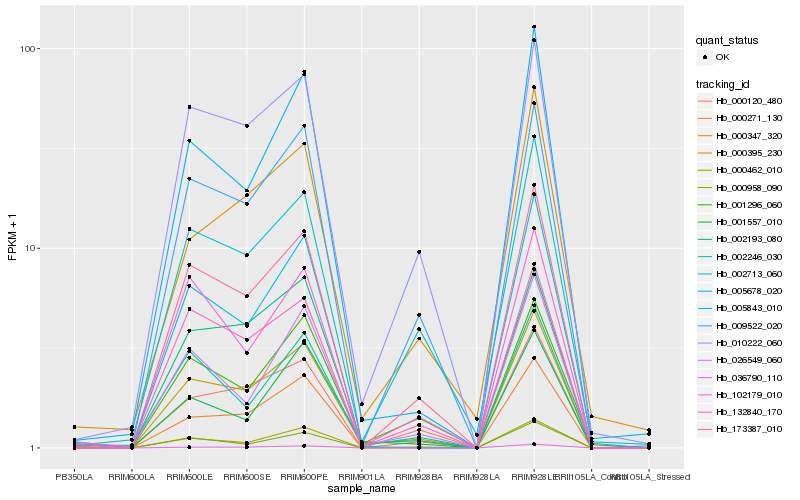
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000395_230 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Vitis vinifera] |
| 2 |
Hb_000271_130 |
0.0827127292 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 3 |
Hb_173387_010 |
0.0896307436 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 4 |
Hb_026549_060 |
0.1042731033 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 5 |
Hb_009522_020 |
0.1043195129 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 6 |
Hb_002193_080 |
0.1101161381 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_005843_010 |
0.11060728 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 8 |
Hb_010222_060 |
0.1106723407 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_002713_060 |
0.1141583391 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 10 |
Hb_102179_010 |
0.114550858 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_001557_010 |
0.1148307085 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 12 |
Hb_132840_170 |
0.1157029443 |
transcription factor |
TF Family: NAC |
Transcription factor [Theobroma cacao] |
| 13 |
Hb_000958_090 |
0.1167642841 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 14 |
Hb_005678_020 |
0.1174843211 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000120_480 |
0.1190903652 |
- |
- |
- |
| 16 |
Hb_001296_060 |
0.1196051793 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 17 |
Hb_002246_030 |
0.1200420759 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 18 |
Hb_000347_320 |
0.1267981849 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 19 |
Hb_000462_010 |
0.1296725223 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |
| 20 |
Hb_036790_110 |
0.1297212166 |
- |
- |
hypothetical protein JCGZ_22271 [Jatropha curcas] |