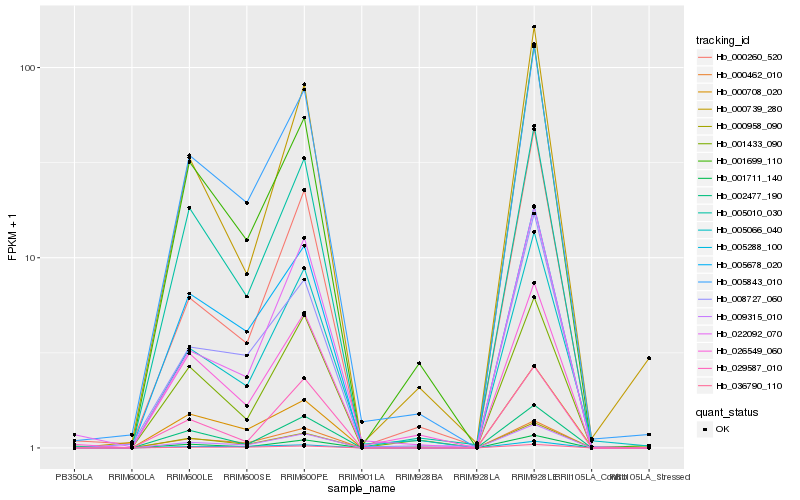
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009315_010 |
0.0 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 2 |
Hb_001711_140 |
0.0567630137 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 3 |
Hb_000462_010 |
0.0600022897 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |
| 4 |
Hb_005843_010 |
0.0636120646 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 5 |
Hb_005678_020 |
0.067696143 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_036790_110 |
0.0776552759 |
- |
- |
hypothetical protein JCGZ_22271 [Jatropha curcas] |
| 7 |
Hb_005010_030 |
0.0806791465 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 8 |
Hb_005066_040 |
0.0827577351 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 9 |
Hb_008727_060 |
0.0859326001 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 10 |
Hb_001699_110 |
0.0945309474 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 11 |
Hb_002477_190 |
0.0957317224 |
- |
- |
hypothetical protein EUGRSUZ_C03380, partial [Eucalyptus grandis] |
| 12 |
Hb_000708_020 |
0.0996393964 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_005288_100 |
0.10025589 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 14 |
Hb_029587_010 |
0.1026816683 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase [Glycine soja] |
| 15 |
Hb_022092_070 |
0.1069444991 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.5 [Jatropha curcas] |
| 16 |
Hb_000739_280 |
0.1091457626 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000260_520 |
0.1100832005 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_001433_090 |
0.1124305819 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 19 |
Hb_026549_060 |
0.1149935979 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 20 |
Hb_000958_090 |
0.1152070048 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |