| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
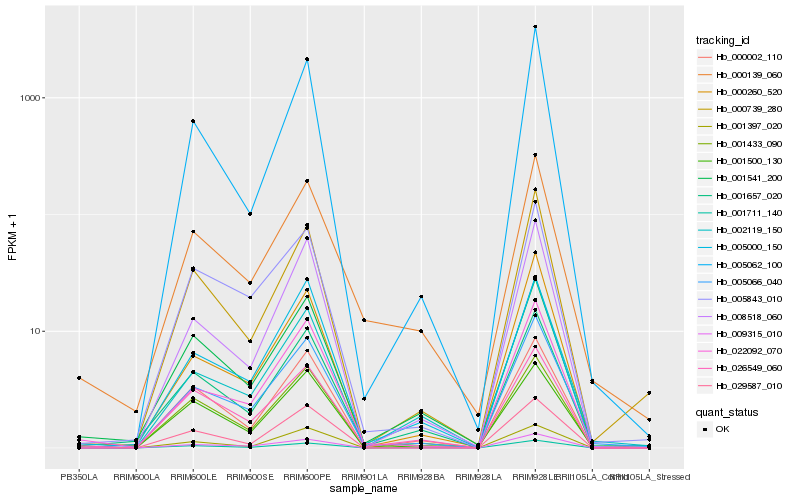
Hb_005066_040 |
0.0 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 2 |
Hb_005843_010 |
0.0792004437 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 3 |
Hb_009315_010 |
0.0827577351 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 4 |
Hb_022092_070 |
0.0851301765 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.5 [Jatropha curcas] |
| 5 |
Hb_002119_150 |
0.085552682 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 6 |
Hb_008518_060 |
0.0917584515 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 7 |
Hb_000739_280 |
0.0920555487 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000260_520 |
0.0940872491 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 9 |
Hb_001711_140 |
0.0944449844 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 10 |
Hb_001657_020 |
0.0970447325 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 11 |
Hb_001433_090 |
0.1001298492 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 12 |
Hb_005062_100 |
0.1009863878 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000002_110 |
0.102490915 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 14 |
Hb_005000_150 |
0.1063263217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000139_060 |
0.1063686565 |
- |
- |
Photosystem I reaction center subunit II, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_001541_200 |
0.1067953688 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 17 |
Hb_026549_060 |
0.107001076 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 18 |
Hb_001500_130 |
0.1071902986 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 19 |
Hb_029587_010 |
0.1082152395 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase [Glycine soja] |
| 20 |
Hb_001397_020 |
0.1085498724 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |