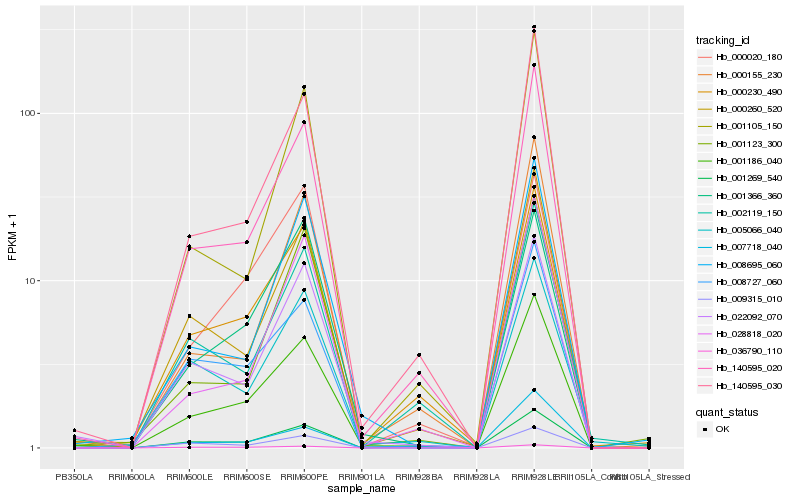
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001186_040 |
0.0 |
- |
- |
Peroxidase 3 precursor, putative [Ricinus communis] |
| 2 |
Hb_140595_020 |
0.0599183733 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Solanum lycopersicum] |
| 3 |
Hb_140595_030 |
0.0816165235 |
- |
- |
- |
| 4 |
Hb_000230_490 |
0.0913912348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_022092_070 |
0.0954209539 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.5 [Jatropha curcas] |
| 6 |
Hb_000260_520 |
0.0957477401 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 7 |
Hb_008727_060 |
0.1025635244 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 8 |
Hb_001269_540 |
0.1094906579 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 9 |
Hb_028818_020 |
0.1164870137 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 10 |
Hb_008695_060 |
0.1174666951 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 11 |
Hb_001366_360 |
0.1204034289 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_005066_040 |
0.1220225241 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 13 |
Hb_000155_230 |
0.1239311949 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 14 |
Hb_001123_300 |
0.124741597 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 15 |
Hb_000020_180 |
0.1255876949 |
- |
- |
laccase, putative [Ricinus communis] |
| 16 |
Hb_007718_040 |
0.1267392852 |
- |
- |
hypothetical protein PRUPE_ppb025054mg [Prunus persica] |
| 17 |
Hb_001105_150 |
0.1270199156 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 18 |
Hb_002119_150 |
0.1282296649 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 19 |
Hb_009315_010 |
0.1289466963 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 20 |
Hb_036790_110 |
0.1309125034 |
- |
- |
hypothetical protein JCGZ_22271 [Jatropha curcas] |